 |
PDBsum entry 1jii

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.1
- tyrosine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Tyr) + L-tyrosine + ATP = L-tyrosyl-tRNA(Tyr) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
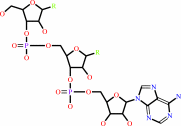 tRNA(Tyr)
tRNA(Tyr)
|
+
|
L-tyrosine
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
 ATP
ATP
|
=
|
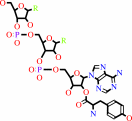 L-tyrosyl-tRNA(Tyr)
L-tyrosyl-tRNA(Tyr)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
10:2008-2016
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
|
|
X.Qiu,
C.A.Janson,
W.W.Smith,
S.M.Green,
P.McDevitt,
K.Johanson,
P.Carter,
M.Hibbs,
C.Lewis,
A.Chalker,
A.Fosberry,
J.Lalonde,
J.Berge,
P.Brown,
C.S.Houge-Frydrych,
R.L.Jarvest.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
SB-219383 and its analogues are a class of potent and specific inhibitors of
bacterial tyrosyl-tRNA synthetases. Crystal structures of these inhibitors have
been solved in complex with the tyrosyl-tRNA synthetase from Staphylococcus
aureus, the bacterium that is largely responsible for hospital-acquired
infections. The full-length enzyme yielded crystals that diffracted to 2.8 A
resolution, but a truncated version of the enzyme allowed the resolution to be
extended to 2.2 A. These inhibitors not only occupy the known substrate binding
sites in unique ways, but also reveal a butyl binding pocket. It was reported
that the Bacillus stearothermophilus TyrRS T51P mutant has much increased
catalytic activity. The S. aureus enzyme happens to have a proline at position
51. Therefore, our structures may contribute to the understanding of the
catalytic mechanism and provide the structural basis for designing novel
antimicrobial agents.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 7.
Fig. 7. The binding of SB-284485 to YRS. (A) Schematic
diagram showing all the hydrogen bonding interactions (dashed
lines) between the inhibitor and YRS. The hydrogen bonding
distances are labeled. (B) Stereoview of the fucose binding
mode. Hydrogen bonds are shown in dashed lines. SB-243545 is
included for comparison. Protein carbons are in yellow,
SB-284485 carbons are in purple, and SB-243545 carbons are in
grey. Oxygen is red and nitrogen is cyan. (C) Stereoview of the
superposition of SB-284485 (purple) and tyrosyl adenylate
(green). The latter is shown with ribose hydrogen bonds and is
modeled based on bsTyrRS structures.
|
 |
Figure 8.
Fig. 8. Stereoviews of the Electron Density (2Fo-Fc) Maps
for the YRS Inhibitors. (A) The density for SB-219383 contoured
at 1  . (B) The
density of SB239629 contoured at 1 . (B) The
density of SB239629 contoured at 1  . (C) The
density for SB-243545 contoured at 1 . (C) The
density for SB-243545 contoured at 1  . (D) The
density for SB-284485 contoured at 1.5 . (D) The
density for SB-284485 contoured at 1.5  . .
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2001,
10,
2008-2016)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Li,
M.Froeyen,
and
P.Herdewijn
(2008).
Comparative structural dynamics of Tyrosyl-tRNA synthetase complexed with different substrates explored by molecular dynamics.
|
| |
Eur Biophys J,
38,
25-35.
|
 |
|
|
|
|
 |
G.Koczyk,
L.S.Wyrwicz,
and
L.Rychlewski
(2007).
LigProf: a simple tool for in silico prediction of ligand-binding sites.
|
| |
J Mol Model,
13,
445-455.
|
 |
|
|
|
|
 |
I.Kufareva,
L.Budagyan,
E.Raush,
M.Totrov,
and
R.Abagyan
(2007).
PIER: protein interface recognition for structural proteomics.
|
| |
Proteins,
67,
400-417.
|
 |
|
|
|
|
 |
L.Bonnefond,
M.Frugier,
E.Touzé,
B.Lorber,
C.Florentz,
R.Giegé,
J.Rudinger-Thirion,
and
C.Sauter
(2007).
Tyrosyl-tRNA synthetase: the first crystallization of a human mitochondrial aminoacyl-tRNA synthetase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
338-341.
|
 |
|
|
|
|
 |
M.Torchala,
and
M.Hoffmann
(2007).
IA, database of known ligands of aminoacyl-tRNA synthetases.
|
| |
J Comput Aided Mol Des,
21,
523-525.
|
 |
|
|
|
|
 |
M.Tsunoda,
Y.Kusakabe,
N.Tanaka,
S.Ohno,
M.Nakamura,
T.Senda,
T.Moriguchi,
N.Asai,
M.Sekine,
T.Yokogawa,
K.Nishikawa,
and
K.T.Nakamura
(2007).
Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
|
| |
Nucleic Acids Res,
35,
4289-4300.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.A.Snyder,
H.J.Feldman,
M.Dumontier,
J.J.Salama,
and
C.W.Hogue
(2006).
Domain-based small molecule binding site annotation.
|
| |
BMC Bioinformatics,
7,
152.
|
 |
|
|
|
|
 |
L.Bonnefond,
M.Frugier,
R.Giegé,
and
J.Rudinger-Thirion
(2005).
Human mitochondrial TyrRS disobeys the tyrosine identity rules.
|
| |
RNA,
11,
558-562.
|
 |
|
|
|
|
 |
T.Kobayashi,
K.Sakamoto,
T.Takimura,
R.Sekine,
V.P.Kelly,
K.Vincent,
K.Kamata,
S.Nishimura,
and
S.Yokoyama
(2005).
Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion.
|
| |
Proc Natl Acad Sci U S A,
102,
1366-1371.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.B.Schmid
(2004).
Seeing is believing: the impact of structural genomics on antimicrobial drug discovery.
|
| |
Nat Rev Microbiol,
2,
739-746.
|
 |
|
|
|
|
 |
T.Kobayashi,
O.Nureki,
R.Ishitani,
A.Yaremchuk,
M.Tukalo,
S.Cusack,
K.Sakamoto,
and
S.Yokoyama
(2003).
Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion.
|
| |
Nat Struct Biol,
10,
425-432.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.L.Yang,
F.J.Otero,
R.J.Skene,
D.E.McRee,
P.Schimmel,
and
L.Ribas de Pouplana
(2003).
Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains.
|
| |
Proc Natl Acad Sci U S A,
100,
15376-15380.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Yaremchuk,
I.Kriklivyi,
M.Tukalo,
and
S.Cusack
(2002).
Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition.
|
| |
EMBO J,
21,
3829-3840.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.L.Yang,
R.J.Skene,
D.E.McRee,
and
P.Schimmel
(2002).
Crystal structure of a human aminoacyl-tRNA synthetase cytokine.
|
| |
Proc Natl Acad Sci U S A,
99,
15369-15374.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

