 |
PDBsum entry 1vbn

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Escherichia coli tyrosyl-tRNA synthetase mutant complexed with tyr-ams
|
|
Structure:
|
 |
Tyrosyl-tRNA synthetase. Chain: a, b. Fragment: residues 5-322. Synonym: tyrosine--tRNA ligase, tyrrs. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.217
|
R-free:
|
0.283
|
|
|
Authors:
|
 |
T.Kobayashi,K.Sakamoto,T.Takimura,K.Kamata,R.Sekine,S.Nishimura, S.Yokoyama,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
Key ref:

|
 |
T.Kobayashi
et al.
(2005).
Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion.
Proc Natl Acad Sci U S A,
102,
1366-1371.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
27-Feb-04
|
Release date:
|
25-Jan-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0AGJ9
(SYY_ECOLI) -
Tyrosine--tRNA ligase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
424 a.a.
318 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.1
- tyrosine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Tyr) + L-tyrosine + ATP = L-tyrosyl-tRNA(Tyr) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
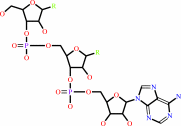 tRNA(Tyr)
tRNA(Tyr)
|
+
|
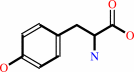 L-tyrosine
L-tyrosine
|
+
|
 ATP
ATP
|
=
|
L-tyrosyl-tRNA(Tyr)
Bound ligand (Het Group name = )
matches with 48.72% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
102:1366-1371
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion.
|
|
T.Kobayashi,
K.Sakamoto,
T.Takimura,
R.Sekine,
V.P.Kelly,
K.Vincent,
K.Kamata,
S.Nishimura,
S.Yokoyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The genetic code in a eukaryotic system has been expanded by the engineering of
Escherichia coli tyrosyl-tRNA synthetase (TyrRS) with the Y37V and Q195C
mutations (37V195C), which specifically recognize 3-iodo-L-tyrosine rather than
L-tyrosine. In the present study, we determined the 3-iodo-L-tyrosine- and
L-tyrosine-bound structures of the 37V195C mutant of the E. coli TyrRS catalytic
domain at 2.0-A resolution. The gamma-methyl group of Val-37 and the sulfur atom
of Cys-195 make van der Waals contacts with the iodine atom of
3-iodo-L-tyrosine. The Val-37 and Cys-195 side chains are rigidly fixed by the
neighboring residues forming the hydrophobic core of the TyrRS. The major roles
of the two mutations are different for the 3-iodo-L-tyrosine-selective
recognition in the first step of the aminoacylation reaction (the amino acid
activation step): the Y37V mutation eliminates the fatal steric repulsion with
the iodine atom, and the Q195C mutation reduces the L-tyrosine misrecognition.
The structure of the 37V195C mutant TyrRS complexed with an L-tyrosyladenylate
analogue was also solved, indicating that the 3-iodo-L-tyrosine and L-tyrosine
side chains are similarly discriminated in the second step (the aminoacyl
transfer step). These results demonstrate that the amino acid-binding pocket on
the 37V195C mutant is optimized for specific 3-iodo-L-tyrosine recognition.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Comparison of the substrate-binding site structures
of E. coli TyrRSs. (A-C) The amino acid-binding site of the
wild-type TyrRS·L-tyrosine (A) (19), 37V195C
TyrRS·3-iodo-L-tyrosine (B), and 37V195C
TyrRS·L-tyrosine (C) complexes. The colors of the atoms
are the same as in Fig. 1 C and D. The hydrogen bonds are
indicated by pink broken lines. (D) Superposed 37V195C (light
blue) and wild-type (green) TyrRS·L-tyrosine structures.
The water molecules are shown by balls. The water molecule that
hydrogen-bonds to the L-tyrosine is labeled "Wat." (E)
Superposed 37V195C mutant TyrRS structures complexed with
3-iodo-L-tyrosine (pink) and L-tyrosine (light blue).
|
 |
Figure 4.
Fig. 4. Superposed 37V195C (pink) and wild-type (light
blue) (19) TyrRS·Tyr-AMS structures. The nitrogen,
oxygen, and sulfur atoms are colored blue, red, and yellow,
respectively.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.Iraha,
K.Oki,
T.Kobayashi,
S.Ohno,
T.Yokogawa,
K.Nishikawa,
S.Yokoyama,
and
K.Sakamoto
(2010).
Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion.
|
| |
Nucleic Acids Res,
38,
3682-3691.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Kawai,
and
S.Yokoyama
(2010).
Professor Tatsuo Miyazawa: from molecular structure to biological function.
|
| |
J Biochem,
148,
631-638.
|
 |
|
|
|
|
 |
K.Sakamoto,
K.Murayama,
K.Oki,
F.Iraha,
M.Kato-Murayama,
M.Takahashi,
K.Ohtake,
T.Kobayashi,
S.Kuramitsu,
M.Shirouzu,
and
S.Yokoyama
(2009).
Genetic encoding of 3-iodo-L-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography.
|
| |
Structure,
17,
335-344.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Brustad,
M.L.Bushey,
J.W.Lee,
D.Groff,
W.Liu,
and
P.G.Schultz
(2008).
A genetically encoded boronate-containing amino acid.
|
| |
Angew Chem Int Ed Engl,
47,
8220-8223.
|
 |
|
|
|
|
 |
K.Oki,
K.Sakamoto,
T.Kobayashi,
H.M.Sasaki,
and
S.Yokoyama
(2008).
Transplantation of a tyrosine editing domain into a tyrosyl-tRNA synthetase variant enhances its specificity for a tyrosine analog.
|
| |
Proc Natl Acad Sci U S A,
105,
13298-13303.
|
 |
|
|
|
|
 |
T.L.Hendrickson
(2008).
Proofreading optimizes iodotyrosine insertion into the genetic code.
|
| |
Proc Natl Acad Sci U S A,
105,
13699-13700.
|
 |
|
|
|
|
 |
M.Tsunoda,
Y.Kusakabe,
N.Tanaka,
S.Ohno,
M.Nakamura,
T.Senda,
T.Moriguchi,
N.Asai,
M.Sekine,
T.Yokogawa,
K.Nishikawa,
and
K.T.Nakamura
(2007).
Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
|
| |
Nucleic Acids Res,
35,
4289-4300.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.A.Cropp,
J.C.Anderson,
and
J.W.Chin
(2007).
Reprogramming the amino-acid substrate specificity of orthogonal aminoacyl-tRNA synthetases to expand the genetic code of eukaryotic cells.
|
| |
Nat Protoc,
2,
2590-2600.
|
 |
|
|
|
|
 |
J.M.Turner,
J.Graziano,
G.Spraggon,
and
P.G.Schultz
(2006).
Structural plasticity of an aminoacyl-tRNA synthetase active site.
|
| |
Proc Natl Acad Sci U S A,
103,
6483-6488.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Wang,
J.Xie,
and
P.G.Schultz
(2006).
Expanding the genetic code.
|
| |
Annu Rev Biophys Biomol Struct,
35,
225-249.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

