 |
PDBsum entry 1h3e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.1
- tyrosine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Tyr) + L-tyrosine + ATP = L-tyrosyl-tRNA(Tyr) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
tRNA(Tyr)
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
L-tyrosine
Bound ligand (Het Group name = )
matches with 92.31% similarity
|
+
|
 ATP
ATP
|
=
|
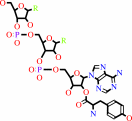 L-tyrosyl-tRNA(Tyr)
L-tyrosyl-tRNA(Tyr)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
21:3829-3840
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition.
|
|
A.Yaremchuk,
I.Kriklivyi,
M.Tukalo,
S.Cusack.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Bacterial tyrosyl-tRNA synthetases (TyrRS) possess a flexibly linked C-terminal
domain of approximately 80 residues, which has hitherto been disordered in
crystal structures of the enzyme. We have determined the structure of Thermus
thermophilus TyrRS at 2.0 A resolution in a crystal form in which the C-terminal
domain is ordered, and confirm that the fold is similar to part of the
C-terminal domain of ribosomal protein S4. We have also determined the structure
at 2.9 A resolution of the complex of T.thermophilus TyrRS with cognate
tRNA(tyr)(G Psi A). In this structure, the C-terminal domain binds between the
characteristic long variable arm of the tRNA and the anti-codon stem, thus
recognizing the unique shape of the tRNA. The anticodon bases have a novel
conformation with A-36 stacked on G-34, and both G-34 and Psi-35 are
base-specifically recognized. The tRNA binds across the two subunits of the
dimeric enzyme and, remarkably, the mode of recognition of the class I TyrRS for
its cognate tRNA resembles that of a class II synthetase in being from the major
groove side of the acceptor stem.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Interactions between tyrosyl-tRNA synthetase and
tRNA^tyr. (A) The C-terminal domain (orange) binds in the elbow
between the long variable arm and the anti-codon stem of the
tRNA (red backbone, green bases). The anti-codon stem loop
interacts with both the C-terminal domain and the  -helical
domain (pink). The tRNA makes no contact with the catalytic
domain of the same subunit (cyan). (B) The unusual conformation
of the anti-codon triplet in which Ade-36 is stacked on Gua-34,
while Psu-35 bulges out. (C) Base-specific interactions of
Asp-259 from the -helical
domain (pink). The tRNA makes no contact with the catalytic
domain of the same subunit (cyan). (B) The unusual conformation
of the anti-codon triplet in which Ade-36 is stacked on Gua-34,
while Psu-35 bulges out. (C) Base-specific interactions of
Asp-259 from the  -helical
domain with Gua-34 and Asp-423 from the C-terminal domain with
Psu-35. -helical
domain with Gua-34 and Asp-423 from the C-terminal domain with
Psu-35.
|
 |
Figure 4.
Figure 4 Structure of tRNAtyr compared with that of tRNA^ser.
(A) Comparison of the secondary structures of T.thermophilus
tRNA^tyr(G  A)
(left) and tRNA^tyr(GGA) (right), highlighting differences,
conserved in other prokaryotic organisms, that determine the
orientation of the long variable arm. tRNA^tyr nucleotides with
only backbone contacts to TyrRSTT are shown in purple, those
with only base contacts are shown in green and those with
backbone and base contacts are shown in orange. (B) Comparison
of the 3D structures of the base of the long variable arm in
T.thermophilus tRNA^tyr and T.thermophilus tRNA^ser (Biou et
al., 1994), based on the structural alignment in (C). In
tRNA^ser, Gua-20B is unpaired and stacks against the first base
pair of the long variable arm, which comprises A45:U48-1 (top).
In tRNA^tyr, U48-1 is unpaired and stacks against the first base
pair of the long variable arm, which comprises A20B:U48−2
(bottom). (C) View looking down the anticodon stem-loop of the
structural alignment of tRNA^tyr (blue) and tRNA^ser (red) based
on superposition of 46 phosphates from the acceptor stem, D- and
T-loops (r.m.s.d. = 1.16 Å). The tRNA cores have a very
similar structure, but the variable arms project at an angle
differing by A)
(left) and tRNA^tyr(GGA) (right), highlighting differences,
conserved in other prokaryotic organisms, that determine the
orientation of the long variable arm. tRNA^tyr nucleotides with
only backbone contacts to TyrRSTT are shown in purple, those
with only base contacts are shown in green and those with
backbone and base contacts are shown in orange. (B) Comparison
of the 3D structures of the base of the long variable arm in
T.thermophilus tRNA^tyr and T.thermophilus tRNA^ser (Biou et
al., 1994), based on the structural alignment in (C). In
tRNA^ser, Gua-20B is unpaired and stacks against the first base
pair of the long variable arm, which comprises A45:U48-1 (top).
In tRNA^tyr, U48-1 is unpaired and stacks against the first base
pair of the long variable arm, which comprises A20B:U48−2
(bottom). (C) View looking down the anticodon stem-loop of the
structural alignment of tRNA^tyr (blue) and tRNA^ser (red) based
on superposition of 46 phosphates from the acceptor stem, D- and
T-loops (r.m.s.d. = 1.16 Å). The tRNA cores have a very
similar structure, but the variable arms project at an angle
differing by  50°. 50°.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2002,
21,
3829-3840)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Palencia,
T.Crépin,
M.T.Vu,
T.L.Lincecum,
S.A.Martinis,
and
S.Cusack
(2012).
Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase.
|
| |
Nat Struct Mol Biol,
19,
677-684.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.G.Riley,
S.Cooper,
P.Hickey,
J.Rudinger-Thirion,
M.McKenzie,
A.Compton,
S.C.Lim,
D.Thorburn,
M.T.Ryan,
R.Giegé,
M.Bahlo,
and
J.Christodoulou
(2010).
Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome.
|
| |
Am J Hum Genet,
87,
52-59.
|
 |
|
|
|
|
 |
X.Dong,
M.Zhou,
C.Zhong,
B.Yang,
N.Shen,
and
J.Ding
(2010).
Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase.
|
| |
Nucleic Acids Res,
38,
1401-1412.
|
 |
|
|
|
|
 |
E.Guth,
M.Farris,
M.Bovee,
and
C.S.Francklyn
(2009).
Asymmetric amino acid activation by class II histidyl-tRNA synthetase from Escherichia coli.
|
| |
J Biol Chem,
284,
20753-20762.
|
 |
|
|
|
|
 |
G.Sharma,
and
E.A.First
(2009).
Thermodynamic Analysis Reveals a Temperature-dependent Change in the Catalytic Mechanism of Bacillus stearothermophilus Tyrosyl-tRNA Synthetase.
|
| |
J Biol Chem,
284,
4179-4190.
|
 |
|
|
|
|
 |
J.H.Kim,
S.J.Park,
K.Y.Lee,
W.S.Son,
N.Y.Sohn,
A.R.Kwon,
and
B.J.Lee
(2009).
Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding.
|
| |
Proteins,
75,
252-257.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.K.Takimoto,
K.L.Adams,
Z.Xiang,
and
L.Wang
(2009).
Improving orthogonal tRNA-synthetase recognition for efficient unnatural amino acid incorporation and application in mammalian cells.
|
| |
Mol Biosyst,
5,
931-934.
|
 |
|
|
|
|
 |
S.Kamijo,
A.Fujii,
K.Onodera,
and
K.Wakabayashi
(2009).
Analyses of conditions for KMSSS loop in tyrosyl-tRNA synthetase by building a mutant library.
|
| |
J Biochem,
146,
241-250.
|
 |
|
|
|
|
 |
K.Oki,
K.Sakamoto,
T.Kobayashi,
H.M.Sasaki,
and
S.Yokoyama
(2008).
Transplantation of a tyrosine editing domain into a tyrosyl-tRNA synthetase variant enhances its specificity for a tyrosine analog.
|
| |
Proc Natl Acad Sci U S A,
105,
13298-13303.
|
 |
|
|
|
|
 |
L.Bonnefond,
C.Florentz,
R.Giegé,
and
J.Rudinger-Thirion
(2008).
Decreased aminoacylation in pathology-related mutants of mitochondrial tRNATyr is associated with structural perturbations in tRNA architecture.
|
| |
RNA,
14,
641-648.
|
 |
|
|
|
|
 |
N.Shen,
M.Zhou,
B.Yang,
Y.Yu,
X.Dong,
and
J.Ding
(2008).
Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states.
|
| |
Nucleic Acids Res,
36,
1288-1299.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.J.Paukstelis,
and
A.M.Lambowitz
(2008).
Identification and evolution of fungal mitochondrial tyrosyl-tRNA synthetases with group I intron splicing activity.
|
| |
Proc Natl Acad Sci U S A,
105,
6010-6015.
|
 |
|
|
|
|
 |
P.J.Paukstelis,
J.H.Chen,
E.Chase,
A.M.Lambowitz,
and
B.L.Golden
(2008).
Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA.
|
| |
Nature,
451,
94-97.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.I.Hauenstein,
Y.M.Hou,
and
J.J.Perona
(2008).
The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity.
|
| |
J Biol Chem,
283,
21997-22006.
|
 |
|
|
|
|
 |
S.N.Rodin,
and
A.S.Rodin
(2008).
On the origin of the genetic code: signatures of its primordial complementarity in tRNAs and aminoacyl-tRNA synthetases.
|
| |
Heredity,
100,
341-355.
|
 |
|
|
|
|
 |
T.Li,
M.Froeyen,
and
P.Herdewijn
(2008).
Comparative structural dynamics of Tyrosyl-tRNA synthetase complexed with different substrates explored by molecular dynamics.
|
| |
Eur Biophys J,
38,
25-35.
|
 |
|
|
|
|
 |
Y.C.Chen,
and
C.Lim
(2008).
Predicting RNA-binding sites from the protein structure based on electrostatics, evolution and geometry.
|
| |
Nucleic Acids Res,
36,
e29.
|
 |
|
|
|
|
 |
C.Abergel,
J.Rudinger-Thirion,
R.Giegé,
and
J.M.Claverie
(2007).
Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS.
|
| |
J Virol,
81,
12406-12417.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Liu,
H.Gamper,
S.Shtivelband,
S.Hauenstein,
J.J.Perona,
and
Y.M.Hou
(2007).
Kinetic quality control of anticodon recognition by a eukaryotic aminoacyl-tRNA synthetase.
|
| |
J Mol Biol,
367,
1063-1078.
|
 |
|
|
|
|
 |
I.A.Vasil'eva,
and
N.A.Moor
(2007).
Interaction of aminoacyl-tRNA synthetases with tRNA: general principles and distinguishing characteristics of the high-molecular-weight substrate recognition.
|
| |
Biochemistry (Mosc),
72,
247-263.
|
 |
|
|
|
|
 |
L.Bonnefond,
M.Frugier,
E.Touzé,
B.Lorber,
C.Florentz,
R.Giegé,
J.Rudinger-Thirion,
and
C.Sauter
(2007).
Tyrosyl-tRNA synthetase: the first crystallization of a human mitochondrial aminoacyl-tRNA synthetase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
338-341.
|
 |
|
|
|
|
 |
M.Delarue
(2007).
An asymmetric underlying rule in the assignment of codons: possible clue to a quick early evolution of the genetic code via successive binary choices.
|
| |
RNA,
13,
161-169.
|
 |
|
|
|
|
 |
M.E.Budiman,
M.H.Knaggs,
J.S.Fetrow,
and
R.W.Alexander
(2007).
Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase.
|
| |
Proteins,
68,
670-689.
|
 |
|
|
|
|
 |
M.Kapustina,
V.Weinreb,
L.Li,
B.Kuhlman,
and
C.W.Carter
(2007).
A conformational transition state accompanies tryptophan activation by B. stearothermophilus tryptophanyl-tRNA synthetase.
|
| |
Structure,
15,
1272-1284.
|
 |
|
|
|
|
 |
M.T.Vu,
and
S.A.Martinis
(2007).
A unique insert of leucyl-tRNA synthetase is required for aminoacylation and not amino acid editing.
|
| |
Biochemistry,
46,
5170-5176.
|
 |
|
|
|
|
 |
M.Tsunoda,
Y.Kusakabe,
N.Tanaka,
S.Ohno,
M.Nakamura,
T.Senda,
T.Moriguchi,
N.Asai,
M.Sekine,
T.Yokogawa,
K.Nishikawa,
and
K.T.Nakamura
(2007).
Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
|
| |
Nucleic Acids Res,
35,
4289-4300.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.W.Lue,
and
S.O.Kelley
(2007).
A single residue in leucyl-tRNA synthetase affecting amino acid specificity and tRNA aminoacylation.
|
| |
Biochemistry,
46,
4466-4472.
|
 |
|
|
|
|
 |
Y.Bessho,
R.Shibata,
S.Sekine,
K.Murayama,
K.Higashijima,
C.Hori-Takemoto,
M.Shirouzu,
S.Kuramitsu,
and
S.Yokoyama
(2007).
Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
|
| |
Proc Natl Acad Sci U S A,
104,
8293-8298.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Pham,
L.Li,
A.Kim,
O.Erdogan,
V.Weinreb,
G.L.Butterfoss,
B.Kuhlman,
and
C.W.Carter
(2007).
A minimal TrpRS catalytic domain supports sense/antisense ancestry of class I and II aminoacyl-tRNA synthetases.
|
| |
Mol Cell,
25,
851-862.
|
 |
|
|
|
|
 |
N.Shen,
L.Guo,
B.Yang,
Y.Jin,
and
J.Ding
(2006).
Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity.
|
| |
Nucleic Acids Res,
34,
3246-3258.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Bilokapic,
T.Maier,
D.Ahel,
I.Gruic-Sovulj,
D.Söll,
I.Weygand-Durasevic,
and
N.Ban
(2006).
Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition.
|
| |
EMBO J,
25,
2498-2509.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.N.Rodin,
and
A.S.Rodin
(2006).
Partitioning of aminoacyl-tRNA synthetases in two classes could have been encoded in a strand-symmetric RNA world.
|
| |
DNA Cell Biol,
25,
617-626.
|
 |
|
|
|
|
 |
X.L.Yang,
F.J.Otero,
K.L.Ewalt,
J.Liu,
M.A.Swairjo,
C.Köhrer,
U.L.RajBhandary,
R.J.Skene,
D.E.McRee,
and
P.Schimmel
(2006).
Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis.
|
| |
EMBO J,
25,
2919-2929.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Matte,
G.V.Louie,
J.Sivaraman,
M.Cygler,
and
S.K.Burley
(2005).
Structure of the pseudouridine synthase RsuA from Haemophilus influenzae.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
350-354.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Lejeune,
N.Delsaux,
B.Charloteaux,
A.Thomas,
and
R.Brasseur
(2005).
Protein-nucleic acid recognition: statistical analysis of atomic interactions and influence of DNA structure.
|
| |
Proteins,
61,
258-271.
|
 |
|
|
|
|
 |
L.Bonnefond,
M.Frugier,
R.Giegé,
and
J.Rudinger-Thirion
(2005).
Human mitochondrial TyrRS disobeys the tyrosine identity rules.
|
| |
RNA,
11,
558-562.
|
 |
|
|
|
|
 |
M.R.Buddha,
and
B.R.Crane
(2005).
Structures of tryptophanyl-tRNA synthetase II from Deinococcus radiodurans bound to ATP and tryptophan. Insight into subunit cooperativity and domain motions linked to catalysis.
|
| |
J Biol Chem,
280,
31965-31973.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Tukalo,
A.Yaremchuk,
R.Fukunaga,
S.Yokoyama,
and
S.Cusack
(2005).
The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation.
|
| |
Nat Struct Mol Biol,
12,
923-930.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.J.Paukstelis,
R.Coon,
L.Madabusi,
J.Nowakowski,
A.Monzingo,
J.Robertus,
and
A.M.Lambowitz
(2005).
A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface.
|
| |
Mol Cell,
17,
417-428.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2005).
Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition.
|
| |
Nat Struct Mol Biol,
12,
915-922.
|
 |
|
|
|
|
 |
T.Kobayashi,
K.Sakamoto,
T.Takimura,
R.Sekine,
V.P.Kelly,
K.Vincent,
K.Kamata,
S.Nishimura,
and
S.Yokoyama
(2005).
Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion.
|
| |
Proc Natl Acad Sci U S A,
102,
1366-1371.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Zhang,
L.Wang,
P.G.Schultz,
and
I.A.Wilson
(2005).
Crystal structures of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine.
|
| |
Protein Sci,
14,
1340-1349.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Korencic,
C.Polycarpo,
I.Weygand-Durasevic,
and
D.Söll
(2004).
Differential modes of transfer RNASer recognition in Methanosarcina barkeri.
|
| |
J Biol Chem,
279,
48780-48786.
|
 |
|
|
|
|
 |
J.Jia,
X.L.Chen,
L.T.Guo,
Y.D.Yu,
J.P.Ding,
and
Y.X.Jin
(2004).
Residues Lys-149 and Glu-153 switch the aminoacylation of tRNA(Trp) in Bacillus subtilis.
|
| |
J Biol Chem,
279,
41960-41965.
|
 |
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2004).
Crystallization and preliminary X-ray crystallographic study of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1916-1918.
|
 |
|
|
|
|
 |
X.Chen,
G.Mohr,
and
A.M.Lambowitz
(2004).
The Neurospora crassa CYT-18 protein C-terminal RNA-binding domain helps stabilize interdomain tertiary interactions in group I introns.
|
| |
RNA,
10,
634-644.
|
 |
|
|
|
|
 |
Y.G.Zheng,
H.Wei,
C.Ling,
F.Martin,
G.Eriani,
and
E.D.Wang
(2004).
Two distinct domains of the beta subunit of Aquifex aeolicus leucyl-tRNA synthetase are involved in tRNA binding as revealed by a three-hybrid selection.
|
| |
Nucleic Acids Res,
32,
3294-3303.
|
 |
|
|
|
|
 |
Y.Kise,
S.W.Lee,
S.G.Park,
S.Fukai,
T.Sengoku,
R.Ishii,
S.Yokoyama,
S.Kim,
and
O.Nureki
(2004).
A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase.
|
| |
Nat Struct Mol Biol,
11,
149-156.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Yu,
Y.Liu,
N.Shen,
X.Xu,
F.Xu,
J.Jia,
Y.Jin,
E.Arnold,
and
J.Ding
(2004).
Crystal structure of human tryptophanyl-tRNA synthetase catalytic fragment: insights into substrate recognition, tRNA binding, and angiogenesis activity.
|
| |
J Biol Chem,
279,
8378-8388.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.R.Ferré-D'Amaré
(2003).
RNA-modifying enzymes.
|
| |
Curr Opin Struct Biol,
13,
49-55.
|
 |
|
|
|
|
 |
L.D.Sherlin,
and
J.J.Perona
(2003).
tRNA-dependent active site assembly in a class I aminoacyl-tRNA synthetase.
|
| |
Structure,
11,
591-603.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Volpon,
C.Lievre,
M.J.Osborne,
S.Gandhi,
P.Iannuzzi,
R.Larocque,
M.Cygler,
K.Gehring,
and
I.Ekiel
(2003).
The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
|
| |
J Bacteriol,
185,
4204-4210.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Francin,
and
M.Mirande
(2003).
Functional dissection of the eukaryotic-specific tRNA-interacting factor of lysyl-tRNA synthetase.
|
| |
J Biol Chem,
278,
1472-1479.
|
 |
|
|
|
|
 |
R.Giegé
(2003).
Genetic code expansion.
|
| |
Nat Struct Biol,
10,
414-416.
|
 |
|
|
|
|
 |
T.Kobayashi,
O.Nureki,
R.Ishitani,
A.Yaremchuk,
M.Tukalo,
S.Cusack,
K.Sakamoto,
and
S.Yokoyama
(2003).
Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion.
|
| |
Nat Struct Biol,
10,
425-432.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.L.Yang,
F.J.Otero,
R.J.Skene,
D.E.McRee,
P.Schimmel,
and
L.Ribas de Pouplana
(2003).
Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains.
|
| |
Proc Natl Acad Sci U S A,
100,
15376-15380.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Francklyn,
J.J.Perona,
J.Puetz,
and
Y.M.Hou
(2002).
Aminoacyl-tRNA synthetases: versatile players in the changing theater of translation.
|
| |
RNA,
8,
1363-1372.
|
 |
|
|
|
|
 |
X.L.Yang,
R.J.Skene,
D.E.McRee,
and
P.Schimmel
(2002).
Crystal structure of a human aminoacyl-tRNA synthetase cytokine.
|
| |
Proc Natl Acad Sci U S A,
99,
15369-15374.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

