
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of human tryptophanyl-tRNA synthetase
|
|
Structure:
|
 |
Tryptophanyl-tRNA synthetase. Chain: a, b. Synonym: tryptophan--tRNA ligase, trprs, ifp53, hwrs. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: wars. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.208
|
R-free:
|
0.239
|
|
|
Authors:
|
 |
X.-L.Yang,F.J.Otero,R.J.Skene,D.E.Mcree,L.Ribas De Pouplana, P.Schimmel
|
Key ref:

|
 |
X.L.Yang
et al.
(2003).
Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains.
Proc Natl Acad Sci U S A,
100,
15376-15380.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
16-Oct-03
|
Release date:
|
06-Jan-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.6.1.1.2
- tryptophan--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Trp) + L-tryptophan + ATP = L-tryptophyl-tRNA(Trp) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
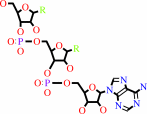 tRNA(Trp)
tRNA(Trp)
|
+
|
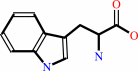 L-tryptophan
L-tryptophan
|
+
|
 ATP
ATP
|
=
|
L-tryptophyl-tRNA(Trp)
Bound ligand (Het Group name = )
matches with 62.16% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
100:15376-15380
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains.
|
|
X.L.Yang,
F.J.Otero,
R.J.Skene,
D.E.McRee,
P.Schimmel,
L.Ribas de Pouplana.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Early forms of the genetic code likely generated "statistical"
proteins, with similar side chains occupying the same sequence positions at
different ratios. In this scenario, groups of related side chains were treated
by aminoacyl-tRNA synthetases as a single molecular species until a
discrimination mechanism developed that could separate them. The aromatic amino
acids tryptophan, tyrosine, and phenylalanine likely constituted one of these
groups. A crystal structure of human tryptophanyl-tRNA synthetase was solved at
2.1 A with a tryptophanyl-adenylate bound at the active site. A cocrystal
structure of an active fragment of human tyrosyl-tRNA synthetase with its
cognate amino acid analog was also solved at 1.6 A. The two structures enabled
active site identifications and provided the information for structure-based
sequence alignments of approximately 45 orthologs of each enzyme. Two critical
positions shared by all tyrosyl-tRNA synthetases and tryptophanyl-tRNA
synthetases for amino acid discrimination were identified. The variations at
these two positions and phylogenetic analyses based on the structural
information suggest that, in contrast to many other amino acids, discrimination
of tyrosine from tryptophan occurred late in the development of the genetic code.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Classification of aminoacyl-tRNA synthetases
adapted from ref. 5. The 20 synthetases are divided into 2
classes of 10 enzymes each. The exceptional class I LysRS is
shown in gray. Highlighted with a yellow box, TyrRS and TrpRS
from class Ic are paired with PheRS from class IIc.
|
 |
Figure 2.
Fig. 2. Structure of the dimeric human TrpRS with one
monomer shown in color. The circled CP1 insertion of the
Rossmann fold domain forms the dimerization interface. All three
domains [N-terminal appended domain (blue), Rossmann fold
catalytic domain (yellow), and anticodon recognition domain
(green)] were resolved in one monomer of the dimer with a
disordered linker of 21 residues connecting the N-domain and the
Rossmann fold domain. However, in the other monomer, the first
96 residues, which include the N-terminal domain, the linker
region, and part of the Rossmann fold catalytic domain, were
completely disordered. A bound Trp-AMP was found only in the
monomer with the resolved N-domain.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Antonczak,
Z.Simova,
I.T.Yonemoto,
M.Bochtler,
A.Piasecka,
H.Czapinska,
A.Brancale,
and
E.M.Tippmann
(2011).
Importance of single molecular determinants in the fidelity of expanded genetic codes.
|
| |
Proc Natl Acad Sci U S A,
108,
1320-1325.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.W.Han,
X.L.Yang,
D.McMullan,
Y.E.Chong,
S.S.Krishna,
C.L.Rife,
D.Weekes,
S.M.Brittain,
P.Abdubek,
E.Ambing,
T.Astakhova,
H.L.Axelrod,
D.Carlton,
J.Caruthers,
H.J.Chiu,
T.Clayton,
L.Duan,
J.Feuerhelm,
J.C.Grant,
S.K.Grzechnik,
L.Jaroszewski,
K.K.Jin,
H.E.Klock,
M.W.Knuth,
A.Kumar,
D.Marciano,
M.D.Miller,
A.T.Morse,
E.Nigoghossian,
L.Okach,
J.Paulsen,
R.Reyes,
H.van den Bedem,
A.White,
G.Wolf,
Q.Xu,
K.O.Hodgson,
J.Wooley,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
M.A.Elsliger,
P.Schimmel,
and
I.A.Wilson
(2010).
Structure of a tryptophanyl-tRNA synthetase containing an iron-sulfur cluster.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1326-1334.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Guo,
P.Schimmel,
and
X.L.Yang
(2010).
Functional expansion of human tRNA synthetases achieved by structural inventions.
|
| |
FEBS Lett,
584,
434-442.
|
 |
|
|
|
|
 |
M.Guo,
X.L.Yang,
and
P.Schimmel
(2010).
New functions of aminoacyl-tRNA synthetases beyond translation.
|
| |
Nat Rev Mol Cell Biol,
11,
668-674.
|
 |
|
|
|
|
 |
M.Zhou,
X.Dong,
N.Shen,
C.Zhong,
and
J.Ding
(2010).
Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design.
|
| |
Nucleic Acids Res,
38,
3399-3413.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Zhou,
M.Kapoor,
M.Guo,
R.Belani,
X.Xu,
W.B.Kiosses,
M.Hanan,
C.Park,
E.Armour,
M.H.Do,
L.A.Nangle,
P.Schimmel,
and
X.L.Yang
(2010).
Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality.
|
| |
Nat Struct Mol Biol,
17,
57-61.
|
 |
|
|
|
|
 |
R.A.Hughes,
and
A.D.Ellington
(2010).
Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA.
|
| |
Nucleic Acids Res,
38,
6813-6830.
|
 |
|
|
|
|
 |
X.Dong,
M.Zhou,
C.Zhong,
B.Yang,
N.Shen,
and
J.Ding
(2010).
Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase.
|
| |
Nucleic Acids Res,
38,
1401-1412.
|
 |
|
|
|
|
 |
F.Charrière,
P.O'Donoghue,
S.Helgadóttir,
L.Maréchal-Drouard,
M.Cristodero,
E.K.Horn,
D.Söll,
and
A.Schneider
(2009).
Dual targeting of a tRNAAsp requires two different aspartyl-tRNA synthetases in Trypanosoma brucei.
|
| |
J Biol Chem,
284,
16210-16217.
|
 |
|
|
|
|
 |
I.A.Vasil'eva,
E.A.Semenova,
and
N.A.Moor
(2009).
Interaction of human phenylalanyl-tRNA synthetase with specific tRNA according to thiophosphate footprinting.
|
| |
Biochemistry (Mosc),
74,
175-185.
|
 |
|
|
|
|
 |
M.Kapoor,
F.J.Otero,
B.M.Slike,
K.L.Ewalt,
and
X.L.Yang
(2009).
Mutational separation of aminoacylation and cytokine activities of human tyrosyl-tRNA synthetase.
|
| |
Chem Biol,
16,
531-539.
|
 |
|
|
|
|
 |
N.Shen,
M.Zhou,
B.Yang,
Y.Yu,
X.Dong,
and
J.Ding
(2008).
Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states.
|
| |
Nucleic Acids Res,
36,
1288-1299.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Abergel,
J.Rudinger-Thirion,
R.Giegé,
and
J.M.Claverie
(2007).
Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS.
|
| |
J Virol,
81,
12406-12417.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Jakó,
P.Ittzés,
A.Szenes,
A.Kun,
E.Szathmáry,
and
G.Pál
(2007).
In silico detection of tRNA sequence features characteristic to aminoacyl-tRNA synthetase class membership.
|
| |
Nucleic Acids Res,
35,
5593-5609.
|
 |
|
|
|
|
 |
G.Koczyk,
L.S.Wyrwicz,
and
L.Rychlewski
(2007).
LigProf: a simple tool for in silico prediction of ligand-binding sites.
|
| |
J Mol Model,
13,
445-455.
|
 |
|
|
|
|
 |
L.T.Guo,
X.L.Chen,
B.T.Zhao,
Y.Shi,
W.Li,
H.Xue,
and
Y.X.Jin
(2007).
Human tryptophanyl-tRNA synthetase is switched to a tRNA-dependent mode for tryptophan activation by mutations at V85 and I311.
|
| |
Nucleic Acids Res,
35,
5934-5943.
|
 |
|
|
|
|
 |
M.Tsunoda,
Y.Kusakabe,
N.Tanaka,
S.Ohno,
M.Nakamura,
T.Senda,
T.Moriguchi,
N.Asai,
M.Sekine,
T.Yokogawa,
K.Nishikawa,
and
K.T.Nakamura
(2007).
Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
|
| |
Nucleic Acids Res,
35,
4289-4300.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Xie,
L.A.Nangle,
W.Zhang,
P.Schimmel,
and
X.L.Yang
(2007).
Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase.
|
| |
Proc Natl Acad Sci U S A,
104,
9976-9981.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.L.Yang,
M.Guo,
M.Kapoor,
K.L.Ewalt,
F.J.Otero,
R.J.Skene,
D.E.McRee,
and
P.Schimmel
(2007).
Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase.
|
| |
Structure,
15,
793-805.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.A.Snyder,
H.J.Feldman,
M.Dumontier,
J.J.Salama,
and
C.W.Hogue
(2006).
Domain-based small molecule binding site annotation.
|
| |
BMC Bioinformatics,
7,
152.
|
 |
|
|
|
|
 |
N.Shen,
L.Guo,
B.Yang,
Y.Jin,
and
J.Ding
(2006).
Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity.
|
| |
Nucleic Acids Res,
34,
3246-3258.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.L.Yang,
F.J.Otero,
K.L.Ewalt,
J.Liu,
M.A.Swairjo,
C.Köhrer,
U.L.RajBhandary,
R.J.Skene,
D.E.McRee,
and
P.Schimmel
(2006).
Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis.
|
| |
EMBO J,
25,
2919-2929.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.R.Buddha,
and
B.R.Crane
(2005).
Structure and activity of an aminoacyl-tRNA synthetase that charges tRNA with nitro-tryptophan.
|
| |
Nat Struct Mol Biol,
12,
274-275.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.R.Buddha,
K.M.Keery,
and
B.R.Crane
(2004).
An unusual tryptophanyl tRNA synthetase interacts with nitric oxide synthase in Deinococcus radiodurans.
|
| |
Proc Natl Acad Sci U S A,
101,
15881-15886.
|
 |
|
|
|
|
 |
P.C.Zamecnik,
M.K.Raychowdhury,
D.R.Tabatadze,
and
H.F.Cantiello
(2004).
Reversal of cystic fibrosis phenotype in a cultured Delta508 cystic fibrosis transmembrane conductance regulator cell line by oligonucleotide insertion.
|
| |
Proc Natl Acad Sci U S A,
101,
8150-8155.
|
 |
|
|
|
|
 |
S.Hauenstein,
C.M.Zhang,
Y.M.Hou,
and
J.J.Perona
(2004).
Shape-selective RNA recognition by cysteinyl-tRNA synthetase.
|
| |
Nat Struct Mol Biol,
11,
1134-1141.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.L.Hendrickson,
V.de Crécy-Lagard,
and
P.Schimmel
(2004).
Incorporation of nonnatural amino acids into proteins.
|
| |
Annu Rev Biochem,
73,
147-176.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

