 |
PDBsum entry 1j4e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Fructose-1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate
|
|
Structure:
|
 |
Fructose-bisphosphate aldolase a. Chain: a, b, c, d. Synonym: muscle-type aldolase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Oryctolagus cuniculus. Rabbit. Organism_taxid: 9986. Tissue: muscle. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.65Å
|
R-factor:
|
0.213
|
R-free:
|
0.249
|
|
|
Authors:
|
 |
K.H.Choi,J.Shi,C.E.Hopkins,D.R.Tolan,K.N.Allen
|
Key ref:

|
 |
K.H.Choi
et al.
(2001).
Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate.
Biochemistry,
40,
13868-13875.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
19-Sep-01
|
Release date:
|
13-Feb-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00883
(ALDOA_RABIT) -
Fructose-bisphosphate aldolase A from Oryctolagus cuniculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
364 a.a.
341 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.13
- fructose-bisphosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
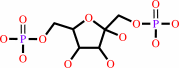 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
=
|
D-glyceraldehyde 3-phosphate
Bound ligand (Het Group name = )
matches with 90.00% similarity
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
40:13868-13875
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate.
|
|
K.H.Choi,
J.Shi,
C.E.Hopkins,
D.R.Tolan,
K.N.Allen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Fructose-1,6-bis(phosphate) aldolase is an essential glycolytic enzyme found in
all vertebrates and higher plants that catalyzes the cleavage of fructose
1,6-bis(phosphate) (Fru-1,6-P(2)) to glyceraldehyde 3-phosphate and
dihydroxyacetone phosphate (DHAP). Mutations in the aldolase genes in humans
cause hemolytic anemia and hereditary fructose intolerance. The structure of the
aldolase-DHAP Schiff base has been determined by X-ray crystallography to 2.6 A
resolution (R(cryst) = 0.213, R(free) = 0.249) by trapping the catalytic
intermediate with NaBH(4) in the presence of Fru-1,6-P(2). This is the first
structure of a trapped covalent intermediate for this essential glycolytic
enzyme. The structure allows the elucidation of a comprehensive catalytic
mechanism and identification of a conserved chemical motif in Schiff-base
aldolases. The position of the bound DHAP relative to Asp33 is consistent with a
role for Asp33 in deprotonation of the C4-hydroxyl leading to C-C bond cleavage.
The methyl side chain of Ala31 is positioned directly opposite the C3-hydroxyl,
sterically favoring the S-configuration of the substrate at this carbon. The
"trigger" residue Arg303, which binds the substrate C6-phosphate group, is a
ligand to the phosphate group of DHAP. The observed movement of the ligand
between substrate and product phosphates may provide a structural link between
the substrate cleavage and the conformational change in the C-terminus
associated with product release. The position of Glu187 in relation to the DHAP
Schiff base is consistent with a role for the residue in protonation of the
hydroxyl group of the carbinolamine in the dehydration step, catalyzing
Schiff-base formation. The overlay of the aldolase-DHAP structure with that of
the covalent enzyme-dihydroxyacetone structure of the mechanistically similar
transaldolase and KDPG aldolase allows the identification of a conserved Lys-Glu
dyad involved in Schiff-base formation and breakdown. The overlay highlights the
fact that Lys146 in aldolase is replaced in transaldolase with Asn35. The
substitution in transaldolase stabilizes the enamine intermediate required for
the attack of the second aldose substrate, changing the chemistry from aldolase
to transaldolase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Rale,
S.Schneider,
G.A.Sprenger,
A.K.Samland,
and
W.D.Fessner
(2011).
Broadening deoxysugar glycodiversity: natural and engineered transaldolases unlock a complementary substrate space.
|
| |
Chemistry,
17,
2623-2632.
|
 |
|
|
|
|
 |
S.Fushinobu,
H.Nishimasu,
D.Hattori,
H.J.Song,
and
T.Wakagi
(2011).
Structural basis for the bifunctionality of fructose-1,6-bisphosphate aldolase/phosphatase.
|
| |
Nature,
478,
538-541.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Esposito,
M.R.Imperato,
L.Ieno,
R.Sorvillo,
V.Benigno,
G.Parenti,
R.Parini,
L.Vitagliano,
A.Zagari,
and
F.Salvatore
(2010).
Hereditary fructose intolerance: functional study of two novel ALDOB natural variants and characterization of a partial gene deletion.
|
| |
Hum Mutat,
31,
1294-1303.
|
 |
|
|
|
|
 |
Z.Diaz,
K.B.Xavier,
and
S.T.Miller
(2009).
The crystal structure of the Escherichia coli autoinducer-2 processing protein LsrF.
|
| |
PLoS One,
4,
e6820.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Sherawat,
D.R.Tolan,
and
K.N.Allen
(2008).
Structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
543-550.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.A.Buscaglia,
W.G.Hol,
V.Nussenzweig,
and
T.Cardozo
(2007).
Modeling the interaction between aldolase and the thrombospondin-related anonymous protein, a key connection of the malaria parasite invasion machinery.
|
| |
Proteins,
66,
528-537.
|
 |
|
|
|
|
 |
J.A.Pezza,
J.D.Stopa,
E.M.Brunyak,
K.N.Allen,
and
D.R.Tolan
(2007).
Thermodynamic analysis shows conformational coupling and dynamics confer substrate specificity in fructose-1,6-bisphosphate aldolase.
|
| |
Biochemistry,
46,
13010-13018.
|
 |
|
|
|
|
 |
J.Bosch,
C.A.Buscaglia,
B.Krumm,
B.P.Ingason,
R.Lucas,
C.Roach,
T.Cardozo,
V.Nussenzweig,
and
W.G.Hol
(2007).
Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
|
| |
Proc Natl Acad Sci U S A,
104,
7015-7020.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.St-Jean,
and
J.Sygusch
(2007).
Stereospecific proton transfer by a mobile catalyst in mammalian fructose-1,6-bisphosphate aldolase.
|
| |
J Biol Chem,
282,
31028-31037.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.St-Jean,
J.Lafrance-Vanasse,
B.Liotard,
and
J.Sygusch
(2005).
High resolution reaction intermediates of rabbit muscle fructose-1,6-bisphosphate aldolase: substrate cleavage and induced fit.
|
| |
J Biol Chem,
280,
27262-27270.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.C.Morais,
G.Zhang,
W.Zhang,
D.B.Olsen,
D.Dunaway-Mariano,
and
K.N.Allen
(2004).
X-ray crystallographic and site-directed mutagenesis analysis of the mechanism of Schiff-base formation in phosphonoacetaldehyde hydrolase catalysis.
|
| |
J Biol Chem,
279,
9353-9361.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.L.Arakaki,
J.A.Pezza,
M.A.Cronin,
C.E.Hopkins,
D.B.Zimmer,
D.R.Tolan,
and
K.N.Allen
(2004).
Structure of human brain fructose 1,6-(bis)phosphate aldolase: linking isozyme structure with function.
|
| |
Protein Sci,
13,
3077-3084.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Lorentzen,
E.Pohl,
P.Zwart,
A.Stark,
R.B.Russell,
T.Knura,
R.Hensel,
and
B.Siebers
(2003).
Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
|
| |
J Biol Chem,
278,
47253-47260.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Pezza,
K.H.Choi,
T.Z.Berardini,
P.T.Beernink,
K.N.Allen,
and
D.R.Tolan
(2003).
Spatial clustering of isozyme-specific residues reveals unlikely determinants of isozyme specificity in fructose-1,6-bisphosphate aldolase.
|
| |
J Biol Chem,
278,
17307-17313.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

