 |
PDBsum entry 1xfb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Human brain fructose 1,6-(bis)phosphate aldolase (c isozyme)
|
|
Structure:
|
 |
AldolasE C. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Synonym: fructose 1,6-(bis)phosphate aldolase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dodecamer (from
)
|
|
Resolution:
|
 |
|
3.00Å
|
R-factor:
|
0.255
|
R-free:
|
0.261
|
|
|
Authors:
|
 |
T.L.Arakaki,J.A.Pezza,M.A.Cronin,C.E.Hopkins,D.B.Zimmer,D.R.Tolan, K.N.Allen
|
Key ref:

|
 |
T.L.Arakaki
et al.
(2004).
Structure of human brain fructose 1,6-(bis)phosphate aldolase: linking isozyme structure with function.
Protein Sci,
13,
3077-3084.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
14-Sep-04
|
Release date:
|
08-Feb-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P09972
(ALDOC_HUMAN) -
Fructose-bisphosphate aldolase C from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
364 a.a.
342 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.13
- fructose-bisphosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
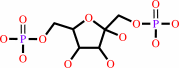 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
13:3077-3084
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of human brain fructose 1,6-(bis)phosphate aldolase: linking isozyme structure with function.
|
|
T.L.Arakaki,
J.A.Pezza,
M.A.Cronin,
C.E.Hopkins,
D.B.Zimmer,
D.R.Tolan,
K.N.Allen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Fructose-1,6-(bis)phosphate aldolase is a ubiquitous enzyme that catalyzes the
reversible aldol cleavage of fructose-1,6-(bis)phosphate and fructose
1-phosphate to dihydroxyacetone phosphate and either glyceral-dehyde-3-phosphate
or glyceraldehyde, respectively. Vertebrate aldolases exist as three isozymes
with different tissue distributions and kinetics: aldolase A (muscle and red
blood cell), aldolase B (liver, kidney, and small intestine), and aldolase C
(brain and neuronal tissue). The structures of human aldolases A and B are known
and herein we report the first structure of the human aldolase C, solved by
X-ray crystallography at 3.0 A resolution. Structural differences between the
isozymes were expected to account for isozyme-specific activity. However, the
structures of isozymes A, B, and C are the same in their overall fold and active
site structure. The subtle changes observed in active site residues Arg42,
Lys146, and Arg303 are insufficient to completely account for the
tissue-specific isozymic differences. Consequently, the structural analysis has
been extended to the isozyme-specific residues (ISRs), those residues conserved
among paralogs. A complete analysis of the ISRs in the context of this structure
demonstrates that in several cases an amino acid residue that is conserved among
aldolase C orthologs prevents an interaction that occurs in paralogs. In
addition, the structure confirms the clustering of ISRs into discrete patches on
the surface and reveals the existence in aldolase C of a patch of
electronegative residues localized near the C terminus. Together, these
structural changes highlight the differences required for the tissue and kinetic
specificity among aldolase isozymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. The active site of aldolase C (red) with residues
depicted as ball-and-stick, overlaid with the active site
residues of aldolase A (blue; PDB accession code 1ALD [PDB]
) and aldolase B (green; PDB accession code 1QO5 [PDB]
).
|
 |
Figure 3.
Figure 3. Stereo view of the active site residues of the
aldolase C structure. The residues are shown as ball-and-stick
models. The 2Fo-Fc electron density map contoured at 1  is depicted
as blue cages. is depicted
as blue cages.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2004,
13,
3077-3084)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.A.Buscaglia,
W.G.Hol,
V.Nussenzweig,
and
T.Cardozo
(2007).
Modeling the interaction between aldolase and the thrombospondin-related anonymous protein, a key connection of the malaria parasite invasion machinery.
|
| |
Proteins,
66,
528-537.
|
 |
|
|
|
|
 |
J.A.Pezza,
J.D.Stopa,
E.M.Brunyak,
K.N.Allen,
and
D.R.Tolan
(2007).
Thermodynamic analysis shows conformational coupling and dynamics confer substrate specificity in fructose-1,6-bisphosphate aldolase.
|
| |
Biochemistry,
46,
13010-13018.
|
 |
|
|
|
|
 |
J.Bosch,
C.A.Buscaglia,
B.Krumm,
B.P.Ingason,
R.Lucas,
C.Roach,
T.Cardozo,
V.Nussenzweig,
and
W.G.Hol
(2007).
Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
|
| |
Proc Natl Acad Sci U S A,
104,
7015-7020.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.R.Gabdoulline,
M.Stein,
and
R.C.Wade
(2007).
qPIPSA: relating enzymatic kinetic parameters and interaction fields.
|
| |
BMC Bioinformatics,
8,
373.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

