 |
PDBsum entry 1zal

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with partially disordered tagatose-1,6-bisphosphate, a weak competitive inhibitor
|
|
Structure:
|
 |
Fructose-bisphosphate aldolase a. Chain: a, b, c, d. Synonym: muscle-type aldolase. Engineered: yes
|
|
Source:
|
 |
Oryctolagus cuniculus. Rabbit. Organism_taxid: 9986. Gene: aldoa. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
1.89Å
|
R-factor:
|
0.168
|
R-free:
|
0.211
|
|
|
Authors:
|
 |
M.St-Jean,J.Lafrance-Vanasse,B.Liotard,J.Sygusch
|
Key ref:

|
 |
M.St-Jean
et al.
(2005).
High resolution reaction intermediates of rabbit muscle fructose-1,6-bisphosphate aldolase: substrate cleavage and induced fit.
J Biol Chem,
280,
27262-27270.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
06-Apr-05
|
Release date:
|
10-May-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00883
(ALDOA_RABIT) -
Fructose-bisphosphate aldolase A from Oryctolagus cuniculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
364 a.a.
363 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.13
- fructose-bisphosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
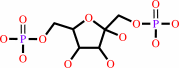 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
=
|
D-glyceraldehyde 3-phosphate
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
280:27262-27270
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
High resolution reaction intermediates of rabbit muscle fructose-1,6-bisphosphate aldolase: substrate cleavage and induced fit.
|
|
M.St-Jean,
J.Lafrance-Vanasse,
B.Liotard,
J.Sygusch.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Crystal structures were determined to 1.8 A resolution of the glycolytic enzyme
fructose-1,6-bis(phosphate) aldolase trapped in complex with its substrate and a
competitive inhibitor, mannitol-1,6-bis(phosphate). The enzyme substrate complex
corresponded to the postulated Schiff base intermediate and has reaction
geometry consistent with incipient C3-C4 bond cleavage catalyzed Glu-187, which
is adjacent by to the Schiff base forming Lys-229. Atom arrangement about the
cleaved bond in the reaction intermediate mimics a pericyclic transition state
occurring in nonenzymatic aldol condensations. Lys-146 hydrogen-bonds the
substrate C4 hydroxyl and assists substrate cleavage by stabilizing the
developing negative charge on the C4 hydroxyl during proton abstraction.
Mannitol-1,6-bis(phosphate) forms a noncovalent complex in the active site whose
binding geometry mimics the covalent carbinolamine precursor. Glu-187
hydrogen-bonds the C2 hydroxyl of the inhibitor in the enzyme complex,
substantiating a proton transfer role by Glu-187 in catalyzing the conversion of
the carbinolamine intermediate to Schiff base. Modeling of the acyclic substrate
configuration into the active site shows Glu-187, in acid form, hydrogen-bonding
both substrate C2 carbonyl and C4 hydroxyl, thereby aligning the substrate
ketose for nucleophilic attack by Lys-229. The multifunctional role of Glu-187
epitomizes a canonical mechanistic feature conserved in Schiff base-forming
aldolases catalyzing carbohydrate metabolism. Trapping of
tagatose-1,6-bis(phosphate), a diastereoisomer of fructose 1,6-bis(phosphate),
displayed stereospecific discrimination and reduced ketohexose binding
specificity. Each ligand induces homologous conformational changes in two
adjacent alpha-helical regions that promote phosphate binding in the active site.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
FIG. 2. Electron density showing the Schiff base
intermediate trapped in the active site of rabbit muscle
aldolase. Difference electron density was calculated from a
1.8-Å annealed F[o] - F[c] omit map encompassing Lys-229
and FBP and contoured at 3  . The green dashes
illustrate hydrogen bonds. A, FBP is covalently bound to the
Schiff base-forming Lys-229 in all subunits, and the FBP O[4] is
hydrogen-bonded to Glu-187 and Lys-146. Orientation is similar
to Fig. 1. B, orientation showing the interaction of active site
residues contacting the Schiff base intermediate. FBP phosphates
interact extensively; the P[1] phosphate makes hydrogen bonding
contacts with Ser-271, Gly-272, Arg-303, and Gly-302, whereas
the P[6] phosphate interacts with Ser-35, Ser-38, and Lys-107.
Orientation differs from Fig. 1 and consists of . The green dashes
illustrate hydrogen bonds. A, FBP is covalently bound to the
Schiff base-forming Lys-229 in all subunits, and the FBP O[4] is
hydrogen-bonded to Glu-187 and Lys-146. Orientation is similar
to Fig. 1. B, orientation showing the interaction of active site
residues contacting the Schiff base intermediate. FBP phosphates
interact extensively; the P[1] phosphate makes hydrogen bonding
contacts with Ser-271, Gly-272, Arg-303, and Gly-302, whereas
the P[6] phosphate interacts with Ser-35, Ser-38, and Lys-107.
Orientation differs from Fig. 1 and consists of  100°
rotation about the 100°
rotation about the  -barrel axis and then
viewing approximately perpendicular to the rotation axis. Some
hydrogen bonds were omitted for visual clarity. -barrel axis and then
viewing approximately perpendicular to the rotation axis. Some
hydrogen bonds were omitted for visual clarity.
|
 |
Figure 4.
FIG. 4. Acyclic form of FBP docked in the active site and
superposition with MBP bound structure. The ketohexose-P[2] was
docked manually by superposition onto the determined MBP
structure, shown in Fig. 3A. The modeled structure was then
subjected to 2000 steps of conjugated gradient minimization with
CNS using topology and parameters from PRODRG. Hydrogen bonding
patterns (green dashes) were conserved when compared with those
in FBP and MBP enzyme adducts. The only significant difference
with respect to the observed enzyme adducts is an additional
hydrogen bond made by Glu-187 with FBP O[2]. The orange dash
illustrates the putative nucleophilic face si attack made on FBP
C[2] carbonyl by Lys-229. Orientation is similar to Fig. 1.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2005,
280,
27262-27270)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Du,
R.F.Say,
W.Lü,
G.Fuchs,
and
O.Einsle
(2011).
Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
|
| |
Nature,
478,
534-537.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Rale,
S.Schneider,
G.A.Sprenger,
A.K.Samland,
and
W.D.Fessner
(2011).
Broadening deoxysugar glycodiversity: natural and engineered transaldolases unlock a complementary substrate space.
|
| |
Chemistry,
17,
2623-2632.
|
 |
|
|
|
|
 |
S.Fushinobu,
H.Nishimasu,
D.Hattori,
H.J.Song,
and
T.Wakagi
(2011).
Structural basis for the bifunctionality of fructose-1,6-bisphosphate aldolase/phosphatase.
|
| |
Nature,
478,
538-541.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.M.Alvarez,
R.García-Rodríguez,
J.M.Martín-Alvarez,
and
D.Miguel
(2010).
Unexpected chemoselectivity in the Schiff condensation of amines with eta(2)(C,O)- eta(1)(O)-coordinated aldehyde.
|
| |
Dalton Trans,
39,
1201-1203.
|
 |
|
|
|
|
 |
G.Esposito,
M.R.Imperato,
L.Ieno,
R.Sorvillo,
V.Benigno,
G.Parenti,
R.Parini,
L.Vitagliano,
A.Zagari,
and
F.Salvatore
(2010).
Hereditary fructose intolerance: functional study of two novel ALDOB natural variants and characterization of a partial gene deletion.
|
| |
Hum Mutat,
31,
1294-1303.
|
 |
|
|
|
|
 |
G.L.Starnes,
M.Coincon,
J.Sygusch,
and
L.D.Sibley
(2009).
Aldolase is essential for energy production and bridging adhesin-actin cytoskeletal interactions during parasite invasion of host cells.
|
| |
Cell Host Microbe,
5,
353-364.
|
 |
|
|
|
|
 |
Y.Sato,
and
M.Nishida
(2009).
Electric charge divergence in proteins: insights into the evolution of their three-dimensional properties.
|
| |
Gene,
441,
3.
|
 |
|
|
|
|
 |
A.Bolt,
A.Berry,
and
A.Nelson
(2008).
Directed evolution of aldolases for exploitation in synthetic organic chemistry.
|
| |
Arch Biochem Biophys,
474,
318-330.
|
 |
|
|
|
|
 |
J.J.Maresh,
L.A.Giddings,
A.Friedrich,
E.A.Loris,
S.Panjikar,
B.L.Trout,
J.Stöckigt,
B.Peters,
and
S.E.O'Connor
(2008).
Strictosidine synthase: mechanism of a Pictet-Spengler catalyzing enzyme.
|
| |
J Am Chem Soc,
130,
710-723.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Sherawat,
D.R.Tolan,
and
K.N.Allen
(2008).
Structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
543-550.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.A.Buscaglia,
W.G.Hol,
V.Nussenzweig,
and
T.Cardozo
(2007).
Modeling the interaction between aldolase and the thrombospondin-related anonymous protein, a key connection of the malaria parasite invasion machinery.
|
| |
Proteins,
66,
528-537.
|
 |
|
|
|
|
 |
J.A.Pezza,
J.D.Stopa,
E.M.Brunyak,
K.N.Allen,
and
D.R.Tolan
(2007).
Thermodynamic analysis shows conformational coupling and dynamics confer substrate specificity in fructose-1,6-bisphosphate aldolase.
|
| |
Biochemistry,
46,
13010-13018.
|
 |
|
|
|
|
 |
J.Bosch,
C.A.Buscaglia,
B.Krumm,
B.P.Ingason,
R.Lucas,
C.Roach,
T.Cardozo,
V.Nussenzweig,
and
W.G.Hol
(2007).
Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
|
| |
Proc Natl Acad Sci U S A,
104,
7015-7020.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Loughman,
and
M.G.Caparon
(2006).
A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes.
|
| |
EMBO J,
25,
5414-5422.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

