 |
PDBsum entry 2pc4

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of fructose-bisphosphate aldolase from plasmodium falciparum in complex with trap-tail determined at 2.4 angstrom resolution
|
|
Structure:
|
 |
Fructose-bisphosphate aldolase. Chain: a, b, c, d. Synonym: 41 kda antigen. Engineered: yes. Pbtrap. Chain: h. Fragment: c-terminus: residues 601-606. Engineered: yes
|
|
Source:
|
 |
Plasmodium falciparum. Malaria parasite p. Falciparum. Organism_taxid: 5833. Expressed in: escherichia coli. Expression_system_taxid: 562. Synthetic: yes. Other_details: synthetic peptide with the sequence based on pbtrap protein from plasmodium berghei, unp entry p90573, p90573_plabe, residues 601-606
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.201
|
R-free:
|
0.250
|
|
|
Authors:
|
 |
J.Bosch,C.A.Buscaglia,B.Krumm,T.Cardozo,V.Nussenzweig,W.G.J.Hol, Structural Genomics Of Pathogenic Protozoa Consortium (Sgpp)
|
Key ref:

|
 |
J.Bosch
et al.
(2007).
Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
Proc Natl Acad Sci U S A,
104,
7015-7020.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
29-Mar-07
|
Release date:
|
17-Apr-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P14223
(ALF_PLAFA) -
Fructose-bisphosphate aldolase from Plasmodium falciparum
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
369 a.a.
359 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.13
- fructose-bisphosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
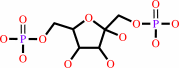 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
104:7015-7020
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
|
|
J.Bosch,
C.A.Buscaglia,
B.Krumm,
B.P.Ingason,
R.Lucas,
C.Roach,
T.Cardozo,
V.Nussenzweig,
W.G.Hol.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
An actomyosin motor located underneath the plasma membrane drives motility and
host-cell invasion of apicomplexan parasites such as Plasmodium falciparum and
Plasmodium vivax, the causative agents of malaria. Aldolase connects the motor
actin filaments to transmembrane adhesive proteins of the thrombospondin-related
anonymous protein (TRAP) family and transduces the motor force across the
parasite surface. The TRAP-aldolase interaction is a distinctive and critical
trait of host hepatocyte invasion by Plasmodium sporozoites, with a likely
similar interaction crucial for erythrocyte invasion by merozoites. Here, we
describe 2.4-A and 2.7-A structures of P. falciparum aldolase (PfAldo) obtained
from crystals grown in the presence of the C-terminal hexapeptide of TRAP from
Plasmodium berghei. The indole ring of the critical penultimate Trp-residue of
TRAP fits snugly into a newly formed hydrophobic pocket, which is exclusively
delimited by hydrophilic residues: two arginines, one glutamate, and one
glutamine. Comparison with the unliganded PfAldo structure shows that the two
arginines adopt new side-chain rotamers, whereas a 25-residue subdomain, forming
a helix-loop-helix unit, shifts upon binding the TRAP-tail. The structural data
are in agreement with decreased TRAP binding after mutagenesis of PfAldo
residues in and near the induced TRAP-binding pocket. Remarkably, the TRAP- and
actin-binding sites of PfAldo seem to overlap, suggesting that both the
plasticity of the aldolase active-site region and the multimeric nature of the
enzyme are crucial for its intriguing nonenzymatic function in the invasion
machinery of the malaria parasite.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Key interactions of the TRAP-tail with P.
falciparum aldolase. (A) TRAP-tail residues D604, W605, and N606
(magenta) interacting with PfAldo residues (light blue). C[  ]positions are marked
as spheres. Selected hydrophobic contacts are shown with red
lines and distances in angstroms. The hydrogen bond between the
indole nitrogen of TRAP-W605 and the carboxylate of E40, and the
interactions between TRAP-N606 with R153 and K151, and of the
backbone oxygen of TRAP-D604 with R48, are depicted in black.
(B) Sequence alignment of the C-terminal TRAP-tail residues of
three plasmodial species. Identical residues are shaded in red.
All TRAP residues involved in contacts with aldolase are well
conserved in several Plasmodium species. ]positions are marked
as spheres. Selected hydrophobic contacts are shown with red
lines and distances in angstroms. The hydrogen bond between the
indole nitrogen of TRAP-W605 and the carboxylate of E40, and the
interactions between TRAP-N606 with R153 and K151, and of the
backbone oxygen of TRAP-D604 with R48, are depicted in black.
(B) Sequence alignment of the C-terminal TRAP-tail residues of
three plasmodial species. Identical residues are shaded in red.
All TRAP residues involved in contacts with aldolase are well
conserved in several Plasmodium species.
|
 |
Figure 4.
Fig. 4. TRAP-tail binding interferes with substrate
binding. Shown is a stereoview of the PfAldo:TRAP-tail complex
superimposed onto the structure of human aldolase A complexed
with F1,6P (30) (PDB ID code 4ALD). Key residues enabling the
accommodation of the penultimate Trp-indole ring are highlighted
as sticks. The TRAP-tail is shown in magenta, residues involved
in substrate binding in blue, and human aldolase A in yellow.
The substrate F1,6P are shown as sticks in green, with the two
phosphate groups in orange. The C-terminal TRAP-tail residue
partially occludes the substrate-binding site.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Kuehn,
N.Simon,
and
G.Pradel
(2010).
Family members stick together: multi-protein complexes of malaria parasites.
|
| |
Med Microbiol Immunol,
199,
209-226.
|
 |
|
|
|
|
 |
E.S.Rangarajan,
H.Park,
E.Fortin,
J.Sygusch,
and
T.Izard
(2010).
Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
|
| |
J Biol Chem,
285,
11983-11990.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.D.Sibley
(2010).
How apicomplexan parasites move in and out of cells.
|
| |
Curr Opin Biotechnol,
21,
592-598.
|
 |
|
|
|
|
 |
P.W.Collingridge,
R.W.Brown,
and
M.L.Ginger
(2010).
Moonlighting enzymes in parasitic protozoa.
|
| |
Parasitology,
137,
1467-1475.
|
 |
|
|
|
|
 |
Y.H.Luah,
B.K.Chaal,
E.Z.Ong,
and
Z.Bozdech
(2010).
A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9?
|
| |
PLoS One,
5,
e10252.
|
 |
|
|
|
|
 |
D.Mazier,
L.Rénia,
and
G.Snounou
(2009).
A pre-emptive strike against malaria's stealthy hepatic forms.
|
| |
Nat Rev Drug Discov,
8,
854-864.
|
 |
|
|
|
|
 |
G.L.Starnes,
M.Coincon,
J.Sygusch,
and
L.D.Sibley
(2009).
Aldolase is essential for energy production and bridging adhesin-actin cytoskeletal interactions during parasite invasion of host cells.
|
| |
Cell Host Microbe,
5,
353-364.
|
 |
|
|
|
|
 |
I.Ejigiri,
and
P.Sinnis
(2009).
Plasmodium sporozoite-host interactions from the dermis to the hepatocyte.
|
| |
Curr Opin Microbiol,
12,
401-407.
|
 |
|
|
|
|
 |
O.Billker,
S.Lourido,
and
L.D.Sibley
(2009).
Calcium-dependent signaling and kinases in apicomplexan parasites.
|
| |
Cell Host Microbe,
5,
612-622.
|
 |
|
|
|
|
 |
W.Daher,
and
D.Soldati-Favre
(2009).
Mechanisms controlling glideosome function in apicomplexans.
|
| |
Curr Opin Microbiol,
12,
408-414.
|
 |
|
|
|
|
 |
F.E.Saenz,
B.Balu,
J.Smith,
S.R.Mendonca,
and
J.H.Adams
(2008).
The transmembrane isoform of Plasmodium falciparum MAEBL is essential for the invasion of Anopheles salivary glands.
|
| |
PLoS ONE,
3,
e2287.
|
 |
|
|
|
|
 |
J.Bosch,
S.Turley,
C.M.Roach,
T.M.Daly,
L.W.Bergman,
and
W.G.Hol
(2007).
The closed MTIP-myosin A-tail complex from the malaria parasite invasion machinery.
|
| |
J Mol Biol,
372,
77-88.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Gayathri,
H.Balaram,
and
M.R.Murthy
(2007).
Structural biology of plasmodial proteins.
|
| |
Curr Opin Struct Biol,
17,
744-754.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

