 |
PDBsum entry 1fro

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lactoylglutathione lyase
|
PDB id
|
|

|

|
1fro
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.4.1.5
- lactoylglutathione lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-S-lactoylglutathione = methylglyoxal + glutathione
|
 |
 |
 |
 |
 |
(R)-S-lactoylglutathione
Bound ligand (Het Group name = )
matches with 79.31% similarity
|
=
|
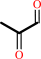 methylglyoxal
methylglyoxal
|
+
|
 glutathione
glutathione
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Embo J
16:3386-3395
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human glyoxalase I--evidence for gene duplication and 3D domain swapping.
|
|
A.D.Cameron,
B.Olin,
M.Ridderström,
B.Mannervik,
T.A.Jones.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The zinc metalloenzyme glyoxalase I catalyses the glutathione-dependent
inactivation of toxic methylglyoxal. The structure of the dimeric human enzyme
in complex with S-benzyl-glutathione has been determined by multiple isomorphous
replacement (MIR) and refined at 2.2 A resolution. Each monomer consists of two
domains. Despite only low sequence homology between them, these domains are
structurally equivalent and appear to have arisen by a gene duplication. On the
other hand, there is no structural homology to the 'glutathione binding domain'
found in other glutathione-linked proteins. 3D domain swapping of the N- and
C-terminal domains has resulted in the active site being situated in the dimer
interface, with the inhibitor and essential zinc ion interacting with side
chains from both subunits. Two structurally equivalent residues from each domain
contribute to a square pyramidal coordination of the zinc ion, rarely seen in
zinc enzymes. Comparison of glyoxalase I with other known structures shows the
enzyme to belong to a new structural family which includes the Fe2+-dependent
dihydroxybiphenyl dioxygenase and the bleomycin resistance protein. This
structural family appears to allow members to form with or without domain
swapping.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Schematic representation of glyoxalase I. (A) Monomer;
(B) dimer. The dimer has been colour ramped according to residue
number, starting with red at the N-terminus of one molecule,
passing through yellow at the C-terminus of that molecule and
finishing with blue at the C-terminus of the other monomer. The
zinc and its coordinating residues are shown in a ball and stick
representation with the zinc coloured green. The active site is
situated in a barrel which is formed only on dimerization.
Residue 114 is situated at the end of the red/yellow domain and
residue 123 at the beginning of the blue/green domain (see the
text). Prepared using MOLSCRIPT (Kraulis, 1991) modified by
R.Esnouf (Oxford University, unpublished). (C) A similar view of
the dihydroxybiphenyl dioxygenase (DHBD) enzyme (Han et al.,
1995) after superposition on the human glyoxalase I enzyme.
Again the molecule has been colour ramped according to residue
number, starting with red at the N-terminus and finishing with
blue at the C-terminus. Despite having only 14% sequence
identity (using the structures to align the sequences), 79 C
 pairs
from the C-terminal domains of this enzyme (blue and green) can
be aligned on glyoxalase I with an r.m.s.d. of 2 Å. The
colouring scheme clearly shows that the suggested domain
swapping in glyoxalase I is not present in DHBD. The ferrous
iron seen in DHBD is situated in a similar position to one of
the zincs in glyoxalase I. The residues coordinating the iron
are structurally equivalent to those binding the zinc. pairs
from the C-terminal domains of this enzyme (blue and green) can
be aligned on glyoxalase I with an r.m.s.d. of 2 Å. The
colouring scheme clearly shows that the suggested domain
swapping in glyoxalase I is not present in DHBD. The ferrous
iron seen in DHBD is situated in a similar position to one of
the zincs in glyoxalase I. The residues coordinating the iron
are structurally equivalent to those binding the zinc.
|
 |
Figure 5.
Figure 5 Proposed reaction mechanism for glyoxalase I. A
shielded base (B) is proposed to abstract the proton from the C1
atom of the hemithioacetal of glutathione and a 2-oxoaldehyde
and then reprotonate at C2.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
Embo J
(1997,
16,
3386-3395)
copyright 1997.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Urscher,
R.Alisch,
and
M.Deponte
(2011).
The glyoxalase system of malaria parasites-Implications for cell biology and general glyoxalase research.
|
| |
Semin Cell Dev Biol,
22,
262-270.
|
 |
|
|
|
|
 |
M.Xue,
N.Rabbani,
and
P.J.Thornalley
(2011).
Glyoxalase in ageing.
|
| |
Semin Cell Dev Biol,
22,
293-301.
|
 |
|
|
|
|
 |
U.Suttisansanee,
and
J.F.Honek
(2011).
Bacterial glyoxalase enzymes.
|
| |
Semin Cell Dev Biol,
22,
285-292.
|
 |
|
|
|
|
 |
C.H.Chu,
W.C.Lo,
H.W.Wang,
Y.C.Hsu,
J.K.Hwang,
P.C.Lyu,
T.W.Pai,
and
C.Y.Tang
(2010).
Detection and alignment of 3D domain swapping proteins using angle-distance image-based secondary structural matching techniques.
|
| |
PLoS One,
5,
e13361.
|
 |
|
|
|
|
 |
C.Q.Scheckhuber,
S.J.Mack,
I.Strobel,
F.Ricciardi,
S.Gispert,
and
H.D.Osiewacz
(2010).
Modulation of the glyoxalase system in the aging model Podospora anserina: effects on growth and lifespan.
|
| |
Aging (Albany NY),
2,
969-980.
|
 |
|
|
|
|
 |
F.Lin,
J.Xu,
J.Shi,
H.Li,
and
B.Li
(2010).
Molecular cloning and characterization of a novel glyoxalase I gene TaGly I in wheat (Triticum aestivum L.).
|
| |
Mol Biol Rep,
37,
729-735.
|
 |
|
|
|
|
 |
G.Birkenmeier,
C.Stegemann,
R.Hoffmann,
R.Günther,
K.Huse,
and
C.Birkemeyer
(2010).
Posttranslational modification of human glyoxalase 1 indicates redox-dependent regulation.
|
| |
PLoS One,
5,
e10399.
|
 |
|
|
|
|
 |
K.Shameer,
G.Pugalenthi,
K.K.Kandaswamy,
P.N.Suganthan,
G.Archunan,
and
R.Sowdhamini
(2010).
Insights into Protein Sequence and Structure-Derived Features Mediating 3D Domain Swapping Mechanism using Support Vector Machine Based Approach.
|
| |
Bioinform Biol Insights,
4,
33-42.
|
 |
|
|
|
|
 |
M.Morar,
and
G.D.Wright
(2010).
The genomic enzymology of antibiotic resistance.
|
| |
Annu Rev Genet,
44,
25-51.
|
 |
|
|
|
|
 |
H.Xu,
Y.Zhang,
J.Yang,
T.Mahmud,
L.Bai,
and
Z.Deng
(2009).
Alternative epimerization in C(7)N-aminocyclitol biosynthesis is catalyzed by ValD, a large protein of the vicinal oxygen chelate superfamily.
|
| |
Chem Biol,
16,
567-576.
|
 |
|
|
|
|
 |
L.Shi,
P.Gao,
X.X.Yan,
and
D.C.Liang
(2009).
Crystal structure of a putative methylmalonyl-coenzyme a epimerase from Thermoanaerobacter tengcongensis at 2.0 A resolution.
|
| |
Proteins,
77,
994-999.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Limphong,
M.W.Crowder,
B.Bennett,
and
C.A.Makaroff
(2009).
Arabidopsis thaliana GLX2-1 contains a dinuclear metal binding site, but is not a glyoxalase 2.
|
| |
Biochem J,
417,
323-330.
|
 |
|
|
|
|
 |
V.de Hemptinne,
D.Rondas,
M.Toepoel,
and
K.Vancompernolle
(2009).
Phosphorylation on Thr-106 and NO-modification of glyoxalase I suppress the TNF-induced transcriptional activity of NF-kappaB.
|
| |
Mol Cell Biochem,
325,
169-178.
|
 |
|
|
|
|
 |
W.Zhang,
L.Wang,
Y.Liu,
J.Xu,
G.Zhu,
H.Cang,
X.Li,
M.Bartlam,
K.Hensley,
G.Li,
Z.Rao,
and
X.C.Zhang
(2009).
Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione.
|
| |
Genes Dev,
23,
1387-1392.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Wu,
P.M.Flatt,
H.Xu,
and
T.Mahmud
(2009).
Biosynthetic Gene Cluster of Cetoniacytone A, an Unusual Aminocyclitol from the Endosymbiotic Bacterium Actinomyces sp. Lu 9419.
|
| |
Chembiochem,
10,
304-314.
|
 |
|
|
|
|
 |
I.Mulako,
J.M.Farrant,
H.Collett,
and
N.Illing
(2008).
Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz.
|
| |
J Exp Bot,
59,
3885-3901.
|
 |
|
|
|
|
 |
M.Kawatani,
H.Okumura,
K.Honda,
N.Kanoh,
M.Muroi,
N.Dohmae,
M.Takami,
M.Kitagawa,
Y.Futamura,
M.Imoto,
and
H.Osada
(2008).
The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I.
|
| |
Proc Natl Acad Sci U S A,
105,
11691-11696.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Deponte,
N.Sturm,
S.Mittler,
M.Harner,
H.Mack,
and
K.Becker
(2007).
Allosteric coupling of two different functional active sites in monomeric Plasmodium falciparum glyoxalase I.
|
| |
J Biol Chem,
282,
28419-28430.
|
 |
|
|
|
|
 |
A.Ariza,
T.J.Vickers,
N.Greig,
K.A.Armour,
M.J.Dixon,
I.M.Eggleston,
A.H.Fairlamb,
and
C.S.Bond
(2006).
Specificity of the trypanothione-dependent Leishmania major glyoxalase I: structure and biochemical comparison with the human enzyme.
|
| |
Mol Microbiol,
59,
1239-1248.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Nocek,
M.Cuff,
E.Evdokimova,
A.Edwards,
A.Joachimiak,
and
A.Savchenko
(2006).
1.6 A crystal structure of a PA2721 protein from pseudomonas aeruginosa--a potential drug-resistance protein.
|
| |
Proteins,
63,
1102-1105.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Ariza,
T.J.Vickers,
N.Greig,
A.H.Fairlamb,
and
C.S.Bond
(2005).
Crystallization and preliminary X-ray analysis of Leishmania major glyoxalase I.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
769-772.
|
 |
|
|
|
|
 |
M.A.Junaid,
D.Kowal,
M.Barua,
P.S.Pullarkat,
S.Sklower Brooks,
and
R.K.Pullarkat
(2004).
Proteomic studies identified a single nucleotide polymorphism in glyoxalase I as autism susceptibility factor.
|
| |
Am J Med Genet A,
131,
11-17.
|
 |
|
|
|
|
 |
M.W.Vetting,
L.P.Wackett,
L.Que,
J.D.Lipscomb,
and
D.H.Ohlendorf
(2004).
Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
|
| |
J Bacteriol,
186,
1945-1958.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Irsch,
and
R.L.Krauth-Siegel
(2004).
Glyoxalase II of African trypanosomes is trypanothione-dependent.
|
| |
J Biol Chem,
279,
22209-22217.
|
 |
|
|
|
|
 |
T.W.Martin,
Z.Dauter,
Y.Devedjiev,
P.Sheffield,
F.Jelen,
M.He,
D.H.Sherman,
J.Otlewski,
Z.S.Derewenda,
and
U.Derewenda
(2002).
Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
|
| |
Structure,
10,
933-942.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Liu,
and
D.Eisenberg
(2002).
3D domain swapping: as domains continue to swap.
|
| |
Protein Sci,
11,
1285-1299.
|
 |
|
|
|
|
 |
A.A.McCarthy,
H.M.Baker,
S.C.Shewry,
M.L.Patchett,
and
E.N.Baker
(2001).
Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold.
|
| |
Structure,
9,
637-646.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Rousseau,
J.W.Schymkowitz,
H.R.Wilkinson,
and
L.S.Itzhaki
(2001).
Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues.
|
| |
Proc Natl Acad Sci U S A,
98,
5596-5601.
|
 |
|
|
|
|
 |
G.Davidson,
S.L.Clugston,
J.F.Honek,
and
M.J.Maroney
(2001).
An XAS investigation of product and inhibitor complexes of Ni-containing GlxI from Escherichia coli: mechanistic implications.
|
| |
Biochemistry,
40,
4569-4582.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
I.S.Mian,
and
I.Dubchak
(2000).
Representing and reasoning about protein families using generative and discriminative methods.
|
| |
J Comput Biol,
7,
849-862.
|
 |
|
|
|
|
 |
K.V.Ramana,
B.L.Dixit,
S.Srivastava,
G.K.Balendiran,
S.K.Srivastava,
and
A.Bhatnagar
(2000).
Selective recognition of glutathiolated aldehydes by aldose reductase.
|
| |
Biochemistry,
39,
12172-12180.
|
 |
|
|
|
|
 |
M.M.He,
S.L.Clugston,
J.F.Honek,
and
B.W.Matthews
(2000).
Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation.
|
| |
Biochemistry,
39,
8719-8727.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.N.Armstrong
(2000).
Mechanistic diversity in a metalloenzyme superfamily.
|
| |
Biochemistry,
39,
13625-13632.
|
 |
|
|
|
|
 |
A.D.Cameron,
M.Ridderström,
B.Olin,
and
B.Mannervik
(1999).
Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
|
| |
Structure,
7,
1067-1078.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.A.Bernat,
L.T.Laughlin,
and
R.N.Armstrong
(1999).
Elucidation of a monovalent cation dependence and characterization of the divalent cation binding site of the fosfomycin resistance protein (FosA).
|
| |
Biochemistry,
38,
7462-7469.
|
 |
|
|
|
|
 |
L.Serre,
A.Sailland,
D.Sy,
P.Boudec,
A.Rolland,
E.Pebay-Peyroula,
and
C.Cohen-Addad
(1999).
Crystal structure of Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase: an enzyme involved in the tyrosine degradation pathway.
|
| |
Structure,
7,
977-988.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Maroney
(1999).
Structure/function relationships in nickel metallobiochemistry.
|
| |
Curr Opin Chem Biol,
3,
188-199.
|
 |
|
|
|
|
 |
Veena,
V.S.Reddy,
and
S.K.Sopory
(1999).
Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress.
|
| |
Plant J,
17,
385-395.
|
 |
|
|
|
|
 |
A.P.Saint-Jean,
K.R.Phillips,
D.J.Creighton,
and
M.J.Stone
(1998).
Active monomeric and dimeric forms of Pseudomonas putida glyoxalase I: evidence for 3D domain swapping.
|
| |
Biochemistry,
37,
10345-10353.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(1998).
Mechanistically diverse enzyme superfamilies: the importance of chemistry in the evolution of catalysis.
|
| |
Curr Opin Chem Biol,
2,
607-612.
|
 |
|
|
|
|
 |
J.M.Dunwell
(1998).
Sequence analysis of the cupin gene family in Synechocystis PCC6803.
|
| |
Microb Comp Genomics,
3,
141-148.
|
 |
|
|
|
|
 |
L.Holm
(1998).
Unification of protein families.
|
| |
Curr Opin Struct Biol,
8,
372-379.
|
 |
|
|
|
|
 |
M.Bergdoll,
L.D.Eltis,
A.D.Cameron,
P.Dumas,
and
J.T.Bolin
(1998).
All in the family: structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly.
|
| |
Protein Sci,
7,
1661-1670.
|
 |
|
|
|
|
 |
M.Ridderström,
A.D.Cameron,
T.A.Jones,
and
B.Mannervik
(1998).
Involvement of an active-site Zn2+ ligand in the catalytic mechanism of human glyoxalase I.
|
| |
J Biol Chem,
273,
21623-21628.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.N.Armstrong
(1998).
Mechanistic imperatives for the evolution of glutathione transferases.
|
| |
Curr Opin Chem Biol,
2,
618-623.
|
 |
|
|
|
|
 |
S.A.Weston,
R.Camble,
J.Colls,
G.Rosenbrock,
I.Taylor,
M.Egerton,
A.D.Tucker,
A.Tunnicliffe,
A.Mistry,
F.Mancia,
E.de la Fortelle,
J.Irwin,
G.Bricogne,
and
R.A.Pauptit
(1998).
Crystal structure of the anti-fungal target N-myristoyl transferase.
|
| |
Nat Struct Biol,
5,
213-221.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.L.Clugston,
J.F.Barnard,
R.Kinach,
D.Miedema,
R.Ruman,
E.Daub,
and
J.F.Honek
(1998).
Overproduction and characterization of a dimeric non-zinc glyoxalase I from Escherichia coli: evidence for optimal activation by nickel ions.
|
| |
Biochemistry,
37,
8754-8763.
|
 |
|
|
|
|
 |
S.Melino,
C.Capo,
B.Dragani,
A.Aceto,
and
R.Petruzzelli
(1998).
A zinc-binding motif conserved in glyoxalase II, beta-lactamase and arylsulfatases.
|
| |
Trends Biochem Sci,
23,
381-382.
|
 |
|
|
|
|
 |
P.C.Babbitt,
and
J.A.Gerlt
(1997).
Understanding enzyme superfamilies. Chemistry As the fundamental determinant in the evolution of new catalytic activities.
|
| |
J Biol Chem,
272,
30591-30594.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

