 |
PDBsum entry 1q0c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1q0c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Anerobic substrate complex of homoprotocatechuate 2,3-dioxygenase from brevibacterium fuscum. (Complex with 3,4-dihydroxyphenylacetate)
|
|
Structure:
|
 |
Homoprotocatechuate 2,3-dioxygenase. Chain: a, b, c, d. Synonym: 3,4-dihydroxyphenylacetate 2,3-dioxygenase. Engineered: yes
|
|
Source:
|
 |
Brevibacterium fuscum. Organism_taxid: 47914. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.170
|
R-free:
|
0.234
|
|
|
Authors:
|
 |
M.W.Vetting,L.P.Wackett,L.Que,J.D.Lipscomb,D.H.Ohlendorf
|
|
Key ref:
|
 |
M.W.Vetting
et al.
(2004).
Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J Bacteriol,
186,
1945-1958.
PubMed id:
|
 |
|
Date:
|
 |
|
15-Jul-03
|
Release date:
|
29-Jul-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q45135
(Q45135_9MICO) -
Homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
365 a.a.
319 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.15
- 3,4-dihydroxyphenylacetate 2,3-dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3,4-dihydroxyphenylacetate + O2 = 2-hydroxy-5-carboxymethylmuconate semialdehyde + H+
|
 |
 |
 |
 |
 |
3,4-dihydroxyphenylacetate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 O2
O2
|
=
|
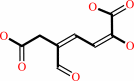 2-hydroxy-5-carboxymethylmuconate semialdehyde
2-hydroxy-5-carboxymethylmuconate semialdehyde
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Bacteriol
186:1945-1958
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
|
|
M.W.Vetting,
L.P.Wackett,
L.Que,
J.D.Lipscomb,
D.H.Ohlendorf.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The X-ray crystal structures of homoprotocatechuate 2,3-dioxygenases isolated
from Arthrobacter globiformis and Brevibacterium fuscum have been determined to
high resolution. These enzymes exhibit 83% sequence identity, yet their
activities depend on different transition metals, Mn2+ and Fe2+, respectively.
The structures allow the origins of metal ion selectivity and aspects of the
molecular mechanism to be examined in detail. The homotetrameric enzymes belong
to the type I family of extradiol dioxygenases (vicinal oxygen chelate
superfamily); each monomer has four betaalphabetabetabeta modules forming two
structurally homologous N-terminal and C-terminal barrel-shaped domains. The
active-site metal is located in the C-terminal barrel and is ligated by two
equatorial ligands, H214NE1 and E267OE1; one axial ligand, H155NE1; and two to
three water molecules. The first and second coordination spheres of these
enzymes are virtually identical (root mean square difference over all atoms,
0.19 A), suggesting that the metal selectivity must be due to changes at a
significant distance from the metal and/or changes that occur during folding.
The substrate (2,3-dihydroxyphenylacetate [HPCA]) chelates the metal
asymmetrically at sites trans to the two imidazole ligands and interacts with a
unique, mobile C-terminal loop. The loop closes over the bound substrate,
presumably to seal the active site as the oxygen activation process commences.
An "open" coordination site trans to E267 is the likely binding site for O2. The
geometry of the enzyme-substrate complexes suggests that if a transiently formed
metal-superoxide complex attacks the substrate without dissociation from the
metal, it must do so at the C-3 position. Second-sphere active-site residues
that are positioned to interact with the HPCA and/or bound O2 during catalysis
are identified and discussed in the context of current mechanistic hypotheses.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.J.Fielding,
E.G.Kovaleva,
E.R.Farquhar,
J.D.Lipscomb,
and
L.Que
(2011).
A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase.
|
| |
J Biol Inorg Chem,
16,
341-355.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.R.Farquhar,
J.P.Emerson,
K.D.Koehntop,
M.F.Reynolds,
M.Trmčić,
and
L.Que
(2011).
In vivo self-hydroxylation of an iron-substituted manganese-dependent extradiol cleaving catechol dioxygenase.
|
| |
J Biol Inorg Chem,
16,
589-597.
|
 |
|
|
|
|
 |
L.M.Blank,
B.E.Ebert,
K.Buehler,
and
B.Bühler
(2010).
Redox biocatalysis and metabolism: molecular mechanisms and metabolic network analysis.
|
| |
Antioxid Redox Signal,
13,
349-394.
|
 |
|
|
|
|
 |
M.M.Mbughuni,
M.Chakrabarti,
J.A.Hayden,
E.L.Bominaar,
M.P.Hendrich,
E.Münck,
and
J.D.Lipscomb
(2010).
Trapping and spectroscopic characterization of an FeIII-superoxo intermediate from a nonheme mononuclear iron-containing enzyme.
|
| |
Proc Natl Acad Sci U S A,
107,
16788-16793.
|
 |
|
|
|
|
 |
H.Suenaga,
S.Mizuta,
and
K.Miyazaki
(2009).
The molecular basis for adaptive evolution in novel extradiol dioxygenases retrieved from the metagenome.
|
| |
FEMS Microbiol Ecol,
69,
472-480.
|
 |
|
|
|
|
 |
J.H.Cho,
D.K.Jung,
K.Lee,
and
S.Rhee
(2009).
Crystal structure and functional analysis of the extradiol dioxygenase LapB from a long-chain alkylphenol degradation pathway in Pseudomonas.
|
| |
J Biol Chem,
284,
34321-34330.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Wagner,
C.Limberg,
and
T.Tietz
(2009).
A novel tripodal ligand containing three different N-heterocyclic donor functions and its application in catechol dioxygenase mimicking.
|
| |
Chemistry,
15,
5567-5576.
|
 |
|
|
|
|
 |
S.Leitgeb,
G.D.Straganz,
and
B.Nidetzky
(2009).
Functional characterization of an orphan cupin protein from Burkholderia xenovorans reveals a mononuclear nonheme Fe2+-dependent oxygenase that cleaves beta-diketones.
|
| |
FEBS J,
276,
5983-5997.
|
 |
|
|
|
|
 |
X.Wu,
P.M.Flatt,
H.Xu,
and
T.Mahmud
(2009).
Biosynthetic Gene Cluster of Cetoniacytone A, an Unusual Aminocyclitol from the Endosymbiotic Bacterium Actinomyces sp. Lu 9419.
|
| |
Chembiochem,
10,
304-314.
|
 |
|
|
|
|
 |
A.F.Miller
(2008).
The shortest wire.
|
| |
Proc Natl Acad Sci U S A,
105,
7341-7342.
|
 |
|
|
|
|
 |
E.G.Kovaleva,
and
J.D.Lipscomb
(2008).
Intermediate in the O-O bond cleavage reaction of an extradiol dioxygenase.
|
| |
Biochemistry,
47,
11168-11170.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Mulako,
J.M.Farrant,
H.Collett,
and
N.Illing
(2008).
Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz.
|
| |
J Exp Bot,
59,
3885-3901.
|
 |
|
|
|
|
 |
J.D.Lipscomb
(2008).
Mechanism of extradiol aromatic ring-cleaving dioxygenases.
|
| |
Curr Opin Struct Biol,
18,
644-649.
|
 |
|
|
|
|
 |
J.P.Emerson,
E.G.Kovaleva,
E.R.Farquhar,
J.D.Lipscomb,
and
L.Que
(2008).
Swapping metals in Fe- and Mn-dependent dioxygenases: evidence for oxygen activation without a change in metal redox state.
|
| |
Proc Natl Acad Sci U S A,
105,
7347-7352.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Moonen,
N.M.Kamerbeek,
A.H.Westphal,
S.A.Boeren,
D.B.Janssen,
M.W.Fraaije,
and
W.J.van Berkel
(2008).
Elucidation of the 4-hydroxyacetophenone catabolic pathway in Pseudomonas fluorescens ACB.
|
| |
J Bacteriol,
190,
5190-5198.
|
 |
|
|
|
|
 |
M.J.Moonen,
S.A.Synowsky,
W.A.van den Berg,
A.H.Westphal,
A.J.Heck,
R.H.van den Heuvel,
M.W.Fraaije,
and
W.J.van Berkel
(2008).
Hydroquinone dioxygenase from pseudomonas fluorescens ACB: a novel member of the family of nonheme-iron(II)-dependent dioxygenases.
|
| |
J Bacteriol,
190,
5199-5209.
|
 |
|
|
|
|
 |
V.Georgiev,
T.Borowski,
M.R.Blomberg,
and
P.E.Siegbahn
(2008).
A comparison of the reaction mechanisms of iron- and manganese-containing 2,3-HPCD: an important spin transition for manganese.
|
| |
J Biol Inorg Chem,
13,
929-940.
|
 |
|
|
|
|
 |
W.A.Gunderson,
A.I.Zatsman,
J.P.Emerson,
E.R.Farquhar,
L.Que,
J.D.Lipscomb,
and
M.P.Hendrich
(2008).
Electron paramagnetic resonance detection of intermediates in the enzymatic cycle of an extradiol dioxygenase.
|
| |
J Am Chem Soc,
130,
14465-14467.
|
 |
|
|
|
|
 |
E.G.Kovaleva,
and
J.D.Lipscomb
(2007).
Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates.
|
| |
Science,
316,
453-457.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.G.Kovaleva,
M.B.Neibergall,
S.Chakrabarty,
and
J.D.Lipscomb
(2007).
Finding intermediates in the O2 activation pathways of non-heme iron oxygenases.
|
| |
Acc Chem Res,
40,
475-483.
|
 |
|
|
|
|
 |
H.Suenaga,
T.Ohnuki,
and
K.Miyazaki
(2007).
Functional screening of a metagenomic library for genes involved in microbial degradation of aromatic compounds.
|
| |
Environ Microbiol,
9,
2289-2297.
|
 |
|
|
|
|
 |
E.F.Mongodin,
N.Shapir,
S.C.Daugherty,
R.T.DeBoy,
J.B.Emerson,
A.Shvartzbeyn,
D.Radune,
J.Vamathevan,
F.Riggs,
V.Grinberg,
H.Khouri,
L.P.Wackett,
K.E.Nelson,
and
M.J.Sadowsky
(2006).
Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1.
|
| |
PLoS Genet,
2,
e214.
|
 |
|
|
|
|
 |
L.Siani,
A.Viggiani,
E.Notomista,
A.Pezzella,
and
A.Di Donato
(2006).
The role of residue Thr249 in modulating the catalytic efficiency and substrate specificity of catechol-2,3-dioxygenase from Pseudomonas stutzeri OX1.
|
| |
FEBS J,
273,
2963-2976.
|
 |
|
|
|
|
 |
S.D.Brown,
J.A.Gerlt,
J.L.Seffernick,
and
P.C.Babbitt
(2006).
A gold standard set of mechanistically diverse enzyme superfamilies.
|
| |
Genome Biol,
7,
R8.
|
 |
|
|
|
|
 |
T.Adachi,
A.Izumi,
D.Rea,
S.Y.Park,
J.R.Tame,
and
D.I.Roper
(2006).
Expression, purification and crystallization of 2-oxo-hept-4-ene-1,7-dioate hydratase (HpcG) from Escherichia coli C.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1010-1012.
|
 |
|
|
|
|
 |
V.Georgiev,
T.Borowski,
and
P.E.Siegbahn
(2006).
Theoretical study of the catalytic reaction mechanism of MndD.
|
| |
J Biol Inorg Chem,
11,
571-585.
|
 |
|
|
|
|
 |
X.Li,
M.Guo,
J.Fan,
W.Tang,
D.Wang,
H.Ge,
H.Rong,
M.Teng,
L.Niu,
Q.Liu,
and
Q.Hao
(2006).
Crystal structure of 3-hydroxyanthranilic acid 3,4-dioxygenase from Saccharomyces cerevisiae: a special subgroup of the type III extradiol dioxygenases.
|
| |
Protein Sci,
15,
761-773.
|
 |
|
|
|
|
 |
J.P.Emerson,
M.L.Wagner,
M.F.Reynolds,
L.Que,
M.J.Sadowsky,
and
L.P.Wackett
(2005).
The role of histidine 200 in MndD, the Mn(II)-dependent 3,4-dihydroxyphenylacetate 2,3-dioxygenase from Arthrobacter globiformis CM-2, a site-directed mutagenesis study.
|
| |
J Biol Inorg Chem,
10,
751-760.
|
 |
|
|
|
|
 |
C.K.Brown,
M.W.Vetting,
C.A.Earhart,
and
D.H.Ohlendorf
(2004).
Biophysical analyses of designed and selected mutants of protocatechuate 3,4-dioxygenase1.
|
| |
Annu Rev Microbiol,
58,
555-585.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Iwata,
H.Noguchi,
Y.Usami,
J.W.Nam,
Z.Fujimoto,
H.Mizuno,
H.Habe,
H.Yamane,
T.Omori,
and
H.Nojiri
(2004).
Crystallization and preliminary crystallographic analysis of the 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase from the carbazole-degrader Pseudomonas resinovorans strain CA10.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2340-2342.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

