 |
PDBsum entry 1fa5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.4.1.5
- lactoylglutathione lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-S-lactoylglutathione = methylglyoxal + glutathione
|
 |
 |
 |
 |
 |
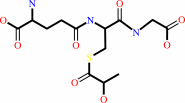 (R)-S-lactoylglutathione
(R)-S-lactoylglutathione
|
=
|
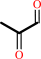 methylglyoxal
methylglyoxal
|
+
|
 glutathione
glutathione
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:8719-8727
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation.
|
|
M.M.He,
S.L.Clugston,
J.F.Honek,
B.W.Matthews.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The metalloenzyme glyoxalase I (GlxI) converts the nonenzymatically produced
hemimercaptal of cytotoxic methylglyoxal and glutathione to nontoxic
S-D-lactoylglutathione. Human GlxI, for which the structure is known, is active
in the presence of Zn(2+). Unexpectedly, the Escherichia coli enzyme is inactive
in the presence of Zn(2+) and is maximally active with Ni(2+). To understand
this difference in metal activation and also to obtain a representative of the
bacterial enzymes, the structure of E. coli Ni(2+)-GlxI has been determined.
Structures have also been determined for the apo enzyme as well as complexes
with Co(2+), Cd(2+), and Zn(2+). It is found that each of the protein-metal
complexes that is catalytically active has octahedral geometry. This includes
the complexes of the E. coli enzyme with Ni(2+), Co(2+), and Cd(2+), as well as
the structures reported for the human Zn(2+) enzyme. Conversely, the complex of
the E. coli enzyme with Zn(2+) has trigonal bipyramidal coordination and is
inactive. This mode of coordination includes four protein ligands plus a single
water molecule. In contrast, the coordination in the active forms of the enzyme
includes two water molecules bound to the metal ion, suggesting that this may be
a key feature of the catalytic mechanism. A comparison of the human and E. coli
enzymes suggests that there are differences between the active sites that might
be exploited for therapeutic use.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
U.Suttisansanee,
and
J.F.Honek
(2011).
Bacterial glyoxalase enzymes.
|
| |
Semin Cell Dev Biol,
22,
285-292.
|
 |
|
|
|
|
 |
K.C.Ryan,
O.E.Johnson,
D.E.Cabelli,
T.C.Brunold,
and
M.J.Maroney
(2010).
Nickel superoxide dismutase: structural and functional roles of Cys2 and Cys6.
|
| |
J Biol Inorg Chem,
15,
795-807.
|
 |
|
|
|
|
 |
L.Shi,
P.Gao,
X.X.Yan,
and
D.C.Liang
(2009).
Crystal structure of a putative methylmalonyl-coenzyme a epimerase from Thermoanaerobacter tengcongensis at 2.0 A resolution.
|
| |
Proteins,
77,
994-999.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.D.Suits,
J.Lang,
G.P.Pal,
M.Couture,
and
Z.Jia
(2009).
Structure and heme binding properties of Escherichia coli O157:H7 ChuX.
|
| |
Protein Sci,
18,
825-838.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.C.Chauhan,
and
R.Madhubala
(2009).
Glyoxalase I gene deletion mutants of Leishmania donovani exhibit reduced methylglyoxal detoxification.
|
| |
PLoS One,
4,
e6805.
|
 |
|
|
|
|
 |
S.W.Ragsdale
(2009).
Nickel-based Enzyme Systems.
|
| |
J Biol Chem,
284,
18571-18575.
|
 |
|
|
|
|
 |
X.Wu,
P.M.Flatt,
H.Xu,
and
T.Mahmud
(2009).
Biosynthetic Gene Cluster of Cetoniacytone A, an Unusual Aminocyclitol from the Endosymbiotic Bacterium Actinomyces sp. Lu 9419.
|
| |
Chembiochem,
10,
304-314.
|
 |
|
|
|
|
 |
N.Sukdeo,
and
J.F.Honek
(2008).
Microbial glyoxalase enzymes: metalloenzymes controlling cellular levels of methylglyoxal.
|
| |
Drug Metabol Drug Interact,
23,
29-50.
|
 |
|
|
|
|
 |
S.K.Yadav,
S.L.Singla-Pareek,
and
S.K.Sopory
(2008).
An overview on the role of methylglyoxal and glyoxalases in plants.
|
| |
Drug Metabol Drug Interact,
23,
51-68.
|
 |
|
|
|
|
 |
M.Deponte,
N.Sturm,
S.Mittler,
M.Harner,
H.Mack,
and
K.Becker
(2007).
Allosteric coupling of two different functional active sites in monomeric Plasmodium falciparum glyoxalase I.
|
| |
J Biol Chem,
282,
28419-28430.
|
 |
|
|
|
|
 |
A.Ariza,
T.J.Vickers,
N.Greig,
K.A.Armour,
M.J.Dixon,
I.M.Eggleston,
A.H.Fairlamb,
and
C.S.Bond
(2006).
Specificity of the trypanothione-dependent Leishmania major glyoxalase I: structure and biochemical comparison with the human enzyme.
|
| |
Mol Microbiol,
59,
1239-1248.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.C.Chen,
J.K.Hwang,
and
J.M.Yang
(2006).
(PS)2: protein structure prediction server.
|
| |
Nucleic Acids Res,
34,
W152-W157.
|
 |
|
|
|
|
 |
A.Ariza,
T.J.Vickers,
N.Greig,
A.H.Fairlamb,
and
C.S.Bond
(2005).
Crystallization and preliminary X-ray analysis of Leishmania major glyoxalase I.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
769-772.
|
 |
|
|
|
|
 |
J.L.Rowe,
G.L.Starnes,
and
P.T.Chivers
(2005).
Complex transcriptional control links NikABCDE-dependent nickel transport with hydrogenase expression in Escherichia coli.
|
| |
J Bacteriol,
187,
6317-6323.
|
 |
|
|
|
|
 |
T.J.Vickers,
N.Greig,
and
A.H.Fairlamb
(2004).
A trypanothione-dependent glyoxalase I with a prokaryotic ancestry in Leishmania major.
|
| |
Proc Natl Acad Sci U S A,
101,
13186-13191.
|
 |
|
|
|
|
 |
S.B.Mulrooney,
and
R.P.Hausinger
(2003).
Nickel uptake and utilization by microorganisms.
|
| |
FEMS Microbiol Rev,
27,
239-261.
|
 |
|
|
|
|
 |
J.S.Cavet,
W.Meng,
M.A.Pennella,
R.J.Appelhoff,
D.P.Giedroc,
and
N.J.Robinson
(2002).
A nickel-cobalt-sensing ArsR-SmtB family repressor. Contributions of cytosol and effector binding sites to metal selectivity.
|
| |
J Biol Chem,
277,
38441-38448.
|
 |
|
|
|
|
 |
P.E.Carrington,
F.Al-Mjeni,
M.A.Zoroddu,
M.Costa,
and
M.J.Maroney
(2002).
Use of XAS for the elucidation of metal structure and function: applications to nickel biochemistry, molecular toxicology, and carcinogenesis.
|
| |
Environ Health Perspect,
110,
705-708.
|
 |
|
|
|
|
 |
T.W.Martin,
Z.Dauter,
Y.Devedjiev,
P.Sheffield,
F.Jelen,
M.He,
D.H.Sherman,
J.Otlewski,
Z.S.Derewenda,
and
U.Derewenda
(2002).
Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
|
| |
Structure,
10,
933-942.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Liu,
and
D.Eisenberg
(2002).
3D domain swapping: as domains continue to swap.
|
| |
Protein Sci,
11,
1285-1299.
|
 |
|
|
|
|
 |
A.A.McCarthy,
H.M.Baker,
S.C.Shewry,
M.L.Patchett,
and
E.N.Baker
(2001).
Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold.
|
| |
Structure,
9,
637-646.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Davidson,
S.L.Clugston,
J.F.Honek,
and
M.J.Maroney
(2001).
An XAS investigation of product and inhibitor complexes of Ni-containing GlxI from Escherichia coli: mechanistic implications.
|
| |
Biochemistry,
40,
4569-4582.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
R.N.Armstrong
(2000).
Mechanistic diversity in a metalloenzyme superfamily.
|
| |
Biochemistry,
39,
13625-13632.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

