 |
PDBsum entry 1fbg

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase(phosphoric monoester)
|
PDB id
|
|

|

|
1fbg
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase(phosphoric monoester)
|
 |
|
Title:
|
 |
Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase
|
|
Structure:
|
 |
Fructose 1,6-bisphosphatase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Sus scrofa. Pig. Organism_taxid: 9823
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
Y.Zhang,J.-Y.Liang,S.Huang,H.Ke,W.N.Lipscomb
|
Key ref:

|
 |
Y.Zhang
et al.
(1993).
Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry,
32,
1844-1857.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
16-Oct-92
|
Release date:
|
31-Oct-93
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00636
(F16P1_PIG) -
Fructose-1,6-bisphosphatase 1 from Sus scrofa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
338 a.a.
313 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.11
- fructose-bisphosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate + H2O = beta-D-fructose 6-phosphate + phosphate
|
 |
 |
 |
 |
 |
beta-D-fructose 1,6-bisphosphate
Bound ligand (Het Group name = )
matches with 95.00% similarity
|
+
|
H2O
|
=
|
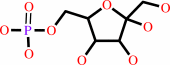 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
32:1844-1857
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
|
|
Y.Zhang,
J.Y.Liang,
S.Huang,
H.Ke,
W.N.Lipscomb.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of fructose-1,6-bisphosphatase (EC 3.1.3.11) complexed
with substrate alone or with substrate analogues in the presence of divalent
metal ions have been determined. The substrate analogues,
2,5-anhydro-D-glucitol-1,6-bisphosphate (AhG-1,6-P2) and
2,5-anhydro-D-mannitol-1,6-bisphosphate (AhM-1,6-P2), differ from the alpha and
beta anomers of fructose-1,6-bisphosphate (Fru-1,6-P2), respectively, in that
the OH on C2 is replaced by a hydrogen atom. Structures have been refined at
resolutions of 2.5 to 3.0 A to R factors of 0.172 to 0.195 with root-mean-square
deviations of 0.012-0.018 A and 2.7-3.8 degrees from the ideal geometries of
bond lengths and bond angles, respectively. In addition, the complex of
substrate with the enzyme has been determined in the absence of metal. The
electron density at 2.5-A resolution does not distinguish between alpha and beta
anomers, which differ for the most part only in the position of the 1-phosphate
group and the orientation of the C2-hydroxyl group. The positions of the
6-phosphate and the sugar ring of the substrate analogues are almost identical
to those of the respective anomer of the substrate. In the presence of metal
ions the positions of the 1-phosphate groups of both alpha and beta analogues
differ significantly (0.8-1.0 A) from those of anomers of the substrate in the
metal-free complex. Two metal ions (Mn2+ or Zn2+) are located at the enzyme
active site of complexes of the alpha analogue AhG-1,6-P2. Metal site 1 is
coordinated by the carboxylate groups of Glu-97, Asp-118, and Glu-280 and the
1-phosphate group of substrate analogue, while the metal site 2 is coordinated
by the carboxylate groups of Glu-97, Asp-118, the 1-phosphate group of substrate
analogue, and the carbonyl oxygen of Leu-120. Both metal sites have a distorted
tetrahedral geometry. However, only one metal ion (Mg2+ or Mn2+) is found very
near the metal site 1 in the enzyme's active site in complexes of the beta
analogue AhM-1,6-P2 or for Mg2+ in the complex of the alpha analogue AhG-1,6-P2.
This single metal ion is coordinated by the carboxylate groups of Glu-97,
Asp-118, Asp-121, and Glu-280 and the 1-phosphate group of substrate analogue in
a distorted square pyramidal geometry.(ABSTRACT TRUNCATED AT 400 WORDS)

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.Li,
K.A.Stieglitz,
A.L.Shrout,
Y.Wei,
R.M.Weis,
B.Stec,
and
M.F.Roberts
(2010).
Mobile loop mutations in an archaeal inositol monophosphatase: modulating three-metal ion assisted catalysis and lithium inhibition.
|
| |
Protein Sci,
19,
309-318.
|
 |
|
|
|
|
 |
A.Burkhardt,
E.T.Spielberg,
S.Simon,
H.Görls,
A.Buchholz,
and
W.Plass
(2009).
Hydrogen Bonds as Structural Directive towards Unusual Polynuclear Complexes: Synthesis, Structure, and Magnetic Properties of Copper(II) and Nickel(II) Complexes with a 2-Aminoglucose Ligand.
|
| |
Chemistry,
15,
1261-1271.
|
 |
|
|
|
|
 |
H.C.Ludwig,
F.N.Pardo,
J.L.Asenjo,
M.A.Maureira,
A.J.Yañez,
and
J.C.Slebe
(2007).
Unraveling multistate unfolding of pig kidney fructose-1,6-bisphosphatase using single tryptophan mutants.
|
| |
FEBS J,
274,
5337-5349.
|
 |
|
|
|
|
 |
R.Gill,
F.Mohammed,
R.Badyal,
L.Coates,
P.Erskine,
D.Thompson,
J.Cooper,
M.Gore,
and
S.Wood
(2005).
High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
545-555.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Cazalis,
A.Chueca,
M.Sahrawy,
and
J.López-Gorgé
(2004).
Construction of chimeric cytosolic fructose-1,6-bisphosphatases by insertion of a chloroplastic redox regulatory cluster.
|
| |
J Physiol Biochem,
60,
7.
|
 |
|
|
|
|
 |
J.Y.Choe,
S.W.Nelson,
H.J.Fromm,
and
R.B.Honzatko
(2003).
Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase.
|
| |
J Biol Chem,
278,
16008-16014.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Teufel,
V.Saudek,
J.P.Ledig,
A.Bernhardt,
S.Boularand,
A.Carreau,
N.J.Cairns,
C.Carter,
D.J.Cowley,
D.Duverger,
A.J.Ganzhorn,
C.Guenet,
B.Heintzelmann,
V.Laucher,
C.Sauvage,
and
T.Smirnova
(2003).
Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase.
|
| |
J Biol Chem,
278,
6521-6531.
|
 |
|
|
|
|
 |
V.Hannaert,
E.Saavedra,
F.Duffieux,
J.P.Szikora,
D.J.Rigden,
P.A.Michels,
and
F.R.Opperdoes
(2003).
Plant-like traits associated with metabolism of Trypanosoma parasites.
|
| |
Proc Natl Acad Sci U S A,
100,
1067-1071.
|
 |
|
|
|
|
 |
N.Kelley-Loughnane,
and
E.R.Kantrowitz
(2001).
Binding of AMP to two of the four subunits of pig kidney fructose-1,6-bisphosphatase induces the allosteric transition.
|
| |
Proteins,
44,
255-261.
|
 |
|
|
|
|
 |
N.Kelley-Loughnane,
and
E.R.Kantrowitz
(2001).
AMP inhibition of pig kidney fructose-1,6-bisphosphatase.
|
| |
Biochim Biophys Acta,
1548,
66-71.
|
 |
|
|
|
|
 |
T.Tanase,
T.Takei,
M.Hidai,
and
S.Yano
(2001).
Substrate-dependent chemoselective aldose-aldose and aldose-ketose isomerizations of carbohydrates promoted by a combination of calcium ion and monoamines.
|
| |
Carbohydr Res,
333,
303-312.
|
 |
|
|
|
|
 |
J.Y.Choe,
H.J.Fromm,
and
R.B.Honzatko
(2000).
Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
|
| |
Biochemistry,
39,
8565-8574.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.W.Nelson,
C.V.Iancu,
J.Y.Choe,
R.B.Honzatko,
and
H.J.Fromm
(2000).
Tryptophan fluorescence reveals the conformational state of a dynamic loop in recombinant porcine fructose-1,6-bisphosphatase.
|
| |
Biochemistry,
39,
11100-11106.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.M.Weeks,
A.W.Roszak,
M.Erman,
R.Kaiser,
H.Jörnvall,
and
D.Ghosh
(1999).
Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
93.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Chiadmi,
A.Navaza,
M.Miginiac-Maslow,
J.P.Jacquot,
and
J.Cherfils
(1999).
Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase.
|
| |
EMBO J,
18,
6809-6815.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.A.Strobel,
and
L.Ortoleva-Donnelly
(1999).
A hydrogen-bonding triad stabilizes the chemical transition state of a group I ribozyme.
|
| |
Chem Biol,
6,
153-165.
|
 |
|
|
|
|
 |
S.A.Strobel
(1999).
A chemogenetic approach to RNA function/structure analysis.
|
| |
Curr Opin Struct Biol,
9,
346-352.
|
 |
|
|
|
|
 |
S.Shan,
A.Yoshida,
S.Sun,
J.A.Piccirilli,
and
D.Herschlag
(1999).
Three metal ions at the active site of the Tetrahymena group I ribozyme.
|
| |
Proc Natl Acad Sci U S A,
96,
12299-12304.
|
 |
|
|
|
|
 |
F.T.Kurbanov,
J.Y.Choe,
R.B.Honzatko,
and
H.J.Fromm
(1998).
Directed mutations in the poorly defined region of porcine liver fructose-1,6-bisphosphatase significantly affect catalysis and the mechanism of AMP inhibition.
|
| |
J Biol Chem,
273,
17511-17516.
|
 |
|
|
|
|
 |
J.E.Coleman
(1998).
Zinc enzymes.
|
| |
Curr Opin Chem Biol,
2,
222-234.
|
 |
|
|
|
|
 |
J.Y.Choe,
B.W.Poland,
H.J.Fromm,
and
R.B.Honzatko
(1998).
Role of a dynamic loop in cation activation and allosteric regulation of recombinant porcine fructose-1,6-bisphosphatase.
|
| |
Biochemistry,
37,
11441-11450.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.J.Hodgson,
Z.Jia,
and
W.C.Plaxton
(1998).
A fluorescence study of ligand-induced conformational changes in cytosolic fructose-1,6-bisphosphatase from germinating castor oil seeds.
|
| |
Biochim Biophys Acta,
1388,
285-294.
|
 |
|
|
|
|
 |
G.J.Narlikar,
and
D.Herschlag
(1997).
Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes.
|
| |
Annu Rev Biochem,
66,
19-59.
|
 |
|
|
|
|
 |
J.R.Atack
(1997).
Inositol monophosphatase inhibitors--lithium mimetics?
|
| |
Med Res Rev,
17,
215-224.
|
 |
|
|
|
|
 |
K.Rees-Milton,
M.Thorne,
P.Greasley,
J.Churchich,
and
M.G.Gore
(1997).
Detection of metal binding to bovine inositol monophosphatase by changes in the near and far ultraviolet regions of the CD spectrum.
|
| |
Eur J Biochem,
246,
211-217.
|
 |
|
|
|
|
 |
L.F.Iversen,
M.Brzozowski,
S.Hastrup,
R.Hubbard,
J.S.Kastrup,
I.K.Larsen,
L.Naerum,
L.Nørskov-Lauridsen,
P.B.Rasmussen,
L.Thim,
F.C.Wiberg,
and
K.Lundgren
(1997).
Characterization of the allosteric binding pocket of human liver fructose-1,6-bisphosphatase by protein crystallography and inhibitor activity studies.
|
| |
Protein Sci,
6,
971-982.
|
 |
|
|
|
|
 |
B.Stec,
R.Abraham,
E.Giroux,
and
E.R.Kantrowitz
(1996).
Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
|
| |
Protein Sci,
5,
1541-1553.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Lu,
B.Stec,
E.L.Giroux,
and
E.R.Kantrowitz
(1996).
Evidence for an active T-state pig kidney fructose 1,6-bisphosphatase: interface residue Lys-42 is important for allosteric inhibition and AMP cooperativity.
|
| |
Protein Sci,
5,
2333-2342.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.F.Shyur,
A.E.Aleshin,
R.B.Honzatko,
and
H.J.Fromm
(1996).
Biochemical properties of mutant and wild-type fructose-1,6-bisphosphatases are consistent with the coupling of intra- and intersubunit conformational changes in the T- and R-state transition.
|
| |
J Biol Chem,
271,
33301-33307.
|
 |
|
|
|
|
 |
R.Zhang,
V.Villeret,
W.N.Lipscomb,
and
H.J.Fromm
(1996).
Kinetics and mechanisms of activation and inhibition of porcine liver fructose-1,6-bisphosphatase by monovalent cations.
|
| |
Biochemistry,
35,
3038-3043.
|
 |
|
|
|
|
 |
G.J.Narlikar,
V.Gopalakrishnan,
T.S.McConnell,
N.Usman,
and
D.Herschlag
(1995).
Use of binding energy by an RNA enzyme for catalysis by positioning and substrate destabilization.
|
| |
Proc Natl Acad Sci U S A,
92,
3668-3672.
|
 |
|
|
|
|
 |
J.P.Jacquot,
J.Lopez-Jaramillo,
A.Chueca,
J.Cherfils,
S.Lemaire,
B.Chedozeau,
M.Miginiac-Maslow,
P.Decottignies,
R.Wolosiuk,
and
J.Lopez-Gorge
(1995).
High-level expression of recombinant pea chloroplast fructose-1,6-bisphosphatase and mutagenesis of its regulatory site.
|
| |
Eur J Biochem,
229,
675-681.
|
 |
|
|
|
|
 |
R.Zhang,
L.Chen,
V.Villeret,
and
H.J.Fromm
(1995).
Glycine 122 is essential for cooperativity and binding of Mg2+ to porcine fructose-1,6-bisphosphatase.
|
| |
J Biol Chem,
270,
54-58.
|
 |
|
|
|
|
 |
V.Villeret,
S.Huang,
H.J.Fromm,
and
W.N.Lipscomb
(1995).
Crystallographic evidence for the action of potassium, thallium, and lithium ions on fructose-1,6-bisphosphatase.
|
| |
Proc Natl Acad Sci U S A,
92,
8916-8920.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Li,
F.J.Stevens,
M.Schiffer,
and
L.E.Anderson
(1994).
Mechanism of light modulation: identification of potential redox-sensitive cysteines distal to catalytic site in light-activated chloroplast enzymes.
|
| |
Biophys J,
67,
29-35.
|
 |
|
|
|
|
 |
P.J.Greasley,
L.G.Hunt,
and
M.G.Gore
(1994).
Bovine inositol monophosphatase. Ligand binding to pyrene-maleimide-labelled enzyme.
|
| |
Eur J Biochem,
222,
453-460.
|
 |
|
|
|
|
 |
S.J.Pollack,
J.R.Atack,
M.R.Knowles,
G.McAllister,
C.I.Ragan,
R.Baker,
S.R.Fletcher,
L.L.Iversen,
and
H.B.Broughton
(1994).
Mechanism of inositol monophosphatase, the putative target of lithium therapy.
|
| |
Proc Natl Acad Sci U S A,
91,
5766-5770.
|
 |
|
|
|
|
 |
Y.Xue,
S.Huang,
J.Y.Liang,
Y.Zhang,
and
W.N.Lipscomb
(1994).
Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
|
| |
Proc Natl Acad Sci U S A,
91,
12482-12486.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Y.Liang,
Y.Zhang,
S.Huang,
and
W.N.Lipscomb
(1993).
Allosteric transition of fructose-1,6-bisphosphatase.
|
| |
Proc Natl Acad Sci U S A,
90,
2132-2136.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

