 |
PDBsum entry 1bfl
|
|

|
| Superseded by: |
 |
|
 |
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Fructose-1,6-bisphosphatase complexed with fructose-6- phosphate and zinc ions
|
|
Structure:
|
 |
Fructose-1,6-bisphosphatase. Chain: a, b. Synonym: fbpase. Engineered: yes. Other_details: the asymmetric unit contains a dimer
|
|
Source:
|
 |
Sus scrofa. Pig. Collection: atcc 00636. Organ: liver. Expressed in: escherichia coli.
|
|
Biol. unit:
|
 |
 Homo-Tetramer (from PDB file)
Homo-Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
2.27Å
|
R-factor:
|
0.190
|
R-free:
|
0.279
|
|
|
Authors:
|
 |
J.Y.Choe,B.W.Poland,H.Fromm,R.Honzatko
|
Key ref:

|
 |
J.Y.Choe
et al.
(1998).
Role of a dynamic loop in cation activation and allosteric regulation of recombinant porcine fructose-1,6-bisphosphatase.
Biochemistry,
37,
11441-11450.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
21-May-98
|
Release date:
|
25-May-99
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00636
(F16P1_PIG) -
Fructose-1,6-bisphosphatase 1 from Sus scrofa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
338 a.a.
324 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.11
- fructose-bisphosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate + H2O = beta-D-fructose 6-phosphate + phosphate
|
 |
 |
 |
 |
 |
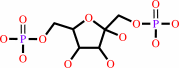 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
+
|
H2O
|
=
|
beta-D-fructose 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:11441-11450
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Role of a dynamic loop in cation activation and allosteric regulation of recombinant porcine fructose-1,6-bisphosphatase.
|
|
J.Y.Choe,
B.W.Poland,
H.J.Fromm,
R.B.Honzatko.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A disordered loop (loop 52-72, residues 52-72) in crystal structures of
fructose-1,6-bisphosphatase (FBPase) has been implicated in regulatory and
catalytic phenomena by studies in directed mutation. A crystal structure of
FBPase in a complex with three zinc cations and the products fructose
6-phosphate (F6P) and phosphate (Pi) reveals loop 52-72 for the first time in a
well-defined conformation with strong electron density. Loop 52-57 interacts
primarily with the active site of its own subunit. Asp68 of the loop hydrogen
bonds with Arg276 and a zinc cation located at the putative potassium activation
site. Leu56 and Tyr57 of the loop pack against hydrophobic residues from two
separate subunits of FBPase. A mechanism of allosteric regulation of catalysis
is presented, in which AMP, by binding to its allosteric pocket, displaces loop
52-72 from the active site. Furthermore, the current structure suggests that
both the alpha- and beta-anomers of F6P can be substrates in the reverse
reaction catalyzed by FBPase. Mechanisms of catalysis are proposed for the
reverse reaction in which Asp121 serves as a catalytic base for the alpha-anomer
and Glu280 serves as a catalytic base for the beta-anomer.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Gizak,
E.Maciaszczyk,
A.Dzugaj,
K.Eschrich,
and
D.Rakus
(2008).
Evolutionary conserved N-terminal region of human muscle fructose 1,6-bisphosphatase regulates its activity and the interaction with aldolase.
|
| |
Proteins,
72,
209-216.
|
 |
|
|
|
|
 |
J.K.Hines,
C.E.Kruesel,
H.J.Fromm,
and
R.B.Honzatko
(2007).
Structure of inhibited fructose-1,6-bisphosphatase from Escherichia coli: distinct allosteric inhibition sites for AMP and glucose 6-phosphate and the characterization of a gluconeogenic switch.
|
| |
J Biol Chem,
282,
24697-24706.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.K.Hines,
X.Chen,
J.C.Nix,
H.J.Fromm,
and
R.B.Honzatko
(2007).
Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition.
|
| |
J Biol Chem,
282,
36121-36131.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.K.Hines,
H.J.Fromm,
and
R.B.Honzatko
(2006).
Novel allosteric activation site in Escherichia coli fructose-1,6-bisphosphatase.
|
| |
J Biol Chem,
281,
18386-18393.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Gill,
F.Mohammed,
R.Badyal,
L.Coates,
P.Erskine,
D.Thompson,
J.Cooper,
M.Gore,
and
S.Wood
(2005).
High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
545-555.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.W.Nelson,
R.B.Honzatko,
and
H.J.Fromm
(2004).
Origin of cooperativity in the activation of fructose-1,6-bisphosphatase by Mg2+.
|
| |
J Biol Chem,
279,
18481-18487.
|
 |
|
|
|
|
 |
D.Rakus,
H.Tillmann,
R.Wysocki,
S.Ulaszewski,
K.Eschrich,
and
A.Dzugaj
(2003).
Different sensitivities of mutants and chimeric forms of human muscle and liver fructose-1,6-bisphosphatases towards AMP.
|
| |
Biol Chem,
384,
51-58.
|
 |
|
|
|
|
 |
J.Y.Choe,
C.V.Iancu,
H.J.Fromm,
and
R.B.Honzatko
(2003).
Metaphosphate in the active site of fructose-1,6-bisphosphatase.
|
| |
J Biol Chem,
278,
16015-16020.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Y.Choe,
S.W.Nelson,
H.J.Fromm,
and
R.B.Honzatko
(2003).
Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase.
|
| |
J Biol Chem,
278,
16008-16014.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Hou,
W.Wang,
H.J.Fromm,
and
R.B.Honzatko
(2002).
IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
|
| |
J Biol Chem,
277,
5970-5976.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Wen,
S.W.Nelson,
R.B.Honzatko,
H.J.Fromm,
and
J.W.Petrich
(2001).
Environment of tryptophan 57 in porcine fructose-1,6-bisphosphatase studied by time-resolved fluorescence and site-directed mutagenesis.
|
| |
Photochem Photobiol,
74,
679-685.
|
 |
|
|
|
|
 |
K.A.Johnson,
L.Chen,
H.Yang,
M.F.Roberts,
and
B.Stec
(2001).
Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
|
| |
Biochemistry,
40,
618-630.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Kelley-Loughnane,
and
E.R.Kantrowitz
(2001).
Binding of AMP to two of the four subunits of pig kidney fructose-1,6-bisphosphatase induces the allosteric transition.
|
| |
Proteins,
44,
255-261.
|
 |
|
|
|
|
 |
N.Kelley-Loughnane,
and
E.R.Kantrowitz
(2001).
AMP inhibition of pig kidney fructose-1,6-bisphosphatase.
|
| |
Biochim Biophys Acta,
1548,
66-71.
|
 |
|
|
|
|
 |
F.W.Zhang,
F.K.Zhao,
and
G.J.Xu
(2000).
Molecular cloning, expression and purification of muscle fructose-1,6-bisphosphatase from Zaocys dhumnades: the role of the N-terminal sequence in AMP activation at alkaline pH.
|
| |
Biol Chem,
381,
561-566.
|
 |
|
|
|
|
 |
J.G.Cárcamo,
A.J.Yañez,
H.C.Ludwig,
O.León,
R.O.Pinto,
A.M.Reyes,
and
J.C.Slebe
(2000).
The C1-C2 interface residue lysine 50 of pig kidney fructose-1, 6-bisphosphatase has a crucial role in the cooperative signal transmission of the AMP inhibition.
|
| |
Eur J Biochem,
267,
2242-2251.
|
 |
|
|
|
|
 |
J.Y.Choe,
H.J.Fromm,
and
R.B.Honzatko
(2000).
Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
|
| |
Biochemistry,
39,
8565-8574.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.W.Nelson,
C.V.Iancu,
J.Y.Choe,
R.B.Honzatko,
and
H.J.Fromm
(2000).
Tryptophan fluorescence reveals the conformational state of a dynamic loop in recombinant porcine fructose-1,6-bisphosphatase.
|
| |
Biochemistry,
39,
11100-11106.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Chiadmi,
A.Navaza,
M.Miginiac-Maslow,
J.P.Jacquot,
and
J.Cherfils
(1999).
Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase.
|
| |
EMBO J,
18,
6809-6815.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

