 |
PDBsum entry 1bk4

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of rabbit liver fructose-1,6-bisphosphatase at 2.3 angstrom resolution
|
|
Structure:
|
 |
Protein (fructose-1,6-bisphosphatase). Chain: a. Synonym: fructose-1,6-diphosphatase. Ec: 3.1.3.11
|
|
Source:
|
 |
Oryctolagus cuniculus. Rabbit. Organism_taxid: 9986. Organ: liver
|
|
Biol. unit:
|
 |
 Tetramer (from PDB file)
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
D.Ghosh,C.M.Weeks,M.Erman,A.W.Roszak,R.Kaiser,H.Jornvall
|
Key ref:

|
 |
C.M.Weeks
et al.
(1999).
Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
Acta Crystallogr D Biol Crystallogr,
55,
93.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
14-Jul-98
|
Release date:
|
22-Jul-98
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00637
(F16P1_RABIT) -
Fructose-1,6-bisphosphatase 1 from Oryctolagus cuniculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
338 a.a.
314 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.11
- fructose-bisphosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate + H2O = beta-D-fructose 6-phosphate + phosphate
|
 |
 |
 |
 |
 |
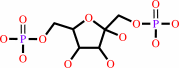 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
+
|
H2O
|
=
|
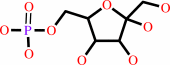 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
55:93
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
|
|
C.M.Weeks,
A.W.Roszak,
M.Erman,
R.Kaiser,
H.Jörnvall,
D.Ghosh.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The three-dimensional structure of the R form of rabbit liver fructose
1,6-bisphosphatase (Fru-1,6-Pase; E.C. 3.1.3.11) has been determined by a
combination of heavy-atom and molecular-replacement methods. A model, which
includes 2394 protein atoms and 86 water molecules, has been refined at 2.3 A
resolution to a crystallographic R factor of 0.177. The root-mean-square
deviations of bond distances and angles from standard geometry are 0.012 A and
1.7 degrees, respectively. This structural result, in conjunction with recently
redetermined amino-acid sequence data, unequivocally establishes that the rabbit
liver enzyme is not an aberrant bisphosphatase as once believed, but is indeed
homologous to other Fru-1,6-Pases. The root-mean-square deviation of the Calpha
atoms in the rabbit liver structure from the homologous atoms in the pig kidney
structure complexed with the product, fructose 6-phosphate, is 0.7 A.
Fru-1,6-Pases are homotetramers, and the rabbit liver protein crystallizes in
space group I222 with one monomer in the asymmetric unit. The structure contains
a single endogenous Mg2+ ion coordinated by Glu97, Asp118, Asp121 and Glu280 at
the site designated metal site 1 in pig kidney Fru-1,6-Pase R-form complexes. In
addition, two sulfate ions, which are found at the positions normally occupied
by the 6-phosphate group of the substrate, as well as the phosphate of the
allosteric inhibitor AMP appear to provide stability. Met177, which has
hydrophobic contacts with the adenine moiety of AMP in pig kidney T-form
complexes, is replaced by glycine. Binding of a non-hydrolyzable substrate
analog, beta-methyl-fructose 1,6-bisphosphate, at the catalytic site is also
examined.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 6.
Figure 6 Coordination of the sulfate ion at the site occupied
by the 6-phosphate group of pig kidney enzyme complexes with
product, competitive inhibitor, substrate and substrate analogs.
Hydrogen-bond distances 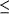 3.2
Å are indicated. 3.2
Å are indicated.
|
 |
Figure 7.
Figure 7 Coordination of the sulfate ion at the site occupied
by the phosphate moiety of AMP in the pig kidney enzyme
complexes. Hydrogen-bond distances 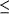 3.2
Å are indicated. 3.2
Å are indicated.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(1999,
55,
93-0)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
 Google scholar
Google scholar
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Berchanski,
B.Shapira,
and
M.Eisenstein
(2004).
Hydrophobic complementarity in protein-protein docking.
|
| |
Proteins,
56,
130-142.
|
 |
|
|
|
|
 |
A.Berchanski,
and
M.Eisenstein
(2003).
Construction of molecular assemblies via docking: modeling of tetramers with D2 symmetry.
|
| |
Proteins,
53,
817-829.
|
 |
|
|
|
|
 |
C.H.Verhees,
J.Akerboom,
E.Schiltz,
W.M.de Vos,
and
J.van der Oost
(2002).
Molecular and biochemical characterization of a distinct type of fructose-1,6-bisphosphatase from Pyrococcus furiosus.
|
| |
J Bacteriol,
184,
3401-3405.
|
 |
|
|
|
|
 |
M.Chiadmi,
A.Navaza,
M.Miginiac-Maslow,
J.P.Jacquot,
and
J.Cherfils
(1999).
Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase.
|
| |
EMBO J,
18,
6809-6815.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

