 |
PDBsum entry 1dwh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.147
- thioglucosidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a thioglucoside + H2O = a sugar + a thiol
|
 |
 |
 |
 |
 |
thioglucoside
Bound ligand (Het Group name = )
matches with 84.62% similarity
|
+
|
H2O
|
=
|
sugar
|
+
|
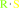 thiol
thiol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
56:328-341
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural changes in a cryo-cooled protein crystal owing to radiation damage.
|
|
W.P.Burmeister.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The high intensity of third-generation X-ray sources, along with the development
of cryo-cooling of protein crystals at temperatures around 100 K, have made it
possible to extend the diffraction limit of crystals and to reduce their size.
However, even with cryo-cooled crystals, radiation damage becomes a limiting
factor. So far, the radiation damage has manifested itself in the form of a loss
of overall diffracted intensity and an increase in the temperature factor. The
structure of a protein (myrosinase) after exposure to different doses of X-rays
in the region of 20 x 10(15) photons mm(-2) has been studied. The changes in the
structure owing to radiation damage were analysed using Fourier difference maps
and occupancy refinement for the first time. Damage was obvious in the form of
breakage of disulfide bonds, decarboxylation of aspartate and glutamate
residues, a loss of hydroxyl groups from tyrosine and of the methylthio group of
methionine. The susceptibility to radiation damage of individual groups of the
same kind varies within the protein. The quality of the model resulting from
structure determination might be compromised owing to the presence of radiolysis
in the crystal after an excessive radiation dose. Radiation-induced structural
changes may interfere with the interpretation of ligand-binding studies or MAD
data. The experiments reported here suggest that there is an intrinsic limit to
the amount of data which can be extracted from a sample of a given size.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Radical reactions caused by X-ray or electron
irradiation which are likely to contribute to the observed
radiation damage to the amino-acid side chains. (p)- represents
protein. References for the reactions are given in the text. The
mechanism for reaction (5) is not known; only the products have
been identified (Schimazu et al., 1964[Berthet-Colominas, C.,
Monaco, S., Novelli, A., Sibaï, G., Mallet, F. & Cusack, S.
(1999). EMBO J. 18, 1124-1136.]).
|
 |
Figure 5.
Figure 5 Individually refined occupancies of labile groups. The
most rapid loss of electron density (plotted with squares,
residue name given) is fitted with an exponential function
(dotted line). The rate constants obtained from these fits are
given in Table 2-; the scatter of the points gives an idea of
the statistical errors of the refined occupancies. The groups
shown in Fig. 3-are plotted as black squares. (a) S atoms of
free cysteines (dashed lines) and disulfide bridges (solid
lines). (b) Carboxyl groups of glutamic acid residues. (c)
Carboxyl groups of aspartate residues. For clarity, the
connecting lines are only drawn for every fifth residue. Asp70
(triangles) is involved in the coordination of the Zn atom. (d)
Hydroxyl groups of tyrosine. For clarity, the connecting lines
are only drawn for every fifth residue. (e) Methylthio groups of
methionine.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2000,
56,
328-341)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.F.White,
N.Noinaj,
Y.Shibata,
J.Love,
B.Kloss,
F.Xu,
J.Gvozdenovic-Jeremic,
P.Shah,
J.Shiloach,
C.G.Tate,
and
R.Grisshammer
(2012).
Structure of the agonist-bound neurotensin receptor.
|
| |
Nature,
490,
508-513.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Royant,
and
M.Noirclerc-Savoye
(2011).
Stabilizing role of glutamic acid 222 in the structure of Enhanced Green Fluorescent Protein.
|
| |
J Struct Biol,
174,
385-390.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Homer,
L.Cooper,
and
A.Gonzalez
(2011).
Energy dependence of site-specific radiation damage in protein crystals.
|
| |
J Synchrotron Radiat,
18,
338-345.
|
 |
|
|
|
|
 |
D.H.Juers,
and
M.Weik
(2011).
Similarities and differences in radiation damage at 100 K versus 160 K in a crystal of thermolysin.
|
| |
J Synchrotron Radiat,
18,
329-337.
|
 |
|
|
|
|
 |
E.De la Mora,
I.Carmichael,
and
E.F.Garman
(2011).
Effective scavenging at cryotemperatures: further increasing the dose tolerance of protein crystals.
|
| |
J Synchrotron Radiat,
18,
346-357.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Karuppasamy,
F.Karimi Nejadasl,
M.Vulovic,
A.J.Koster,
and
R.B.Ravelli
(2011).
Radiation damage in single-particle cryo-electron microscopy: effects of dose and dose rate.
|
| |
J Synchrotron Radiat,
18,
398-412.
|
 |
|
|
|
|
 |
T.Krojer,
and
F.von Delft
(2011).
Assessment of radiation damage behaviour in a large collection of empirically optimized datasets highlights the importance of unmeasured complicating effects.
|
| |
J Synchrotron Radiat,
18,
387-397.
|
 |
|
|
|
|
 |
A.Meents,
S.Gutmann,
A.Wagner,
and
C.Schulze-Briese
(2010).
Origin and temperature dependence of radiation damage in biological samples at cryogenic temperatures.
|
| |
Proc Natl Acad Sci U S A,
107,
1094-1099.
|
 |
|
|
|
|
 |
C.Nicolini,
and
E.Pechkova
(2010).
Nanoproteomics for nanomedicine.
|
| |
Nanomedicine (Lond),
5,
677-682.
|
 |
|
|
|
|
 |
D.Borek,
M.Cymborowski,
M.Machius,
W.Minor,
and
Z.Otwinowski
(2010).
Diffraction data analysis in the presence of radiation damage.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
426-436.
|
 |
|
|
|
|
 |
E.F.Garman
(2010).
Radiation damage in macromolecular crystallography: what is it and why should we care?
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
339-351.
|
 |
|
|
|
|
 |
G.Bujacz,
B.Wrzesniewska,
and
A.Bujacz
(2010).
Cryoprotection properties of salts of organic acids: a case study for a tetragonal crystal of HEW lysozyme.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
789-796.
|
 |
|
|
|
|
 |
G.P.Bourenkov,
and
A.N.Popov
(2010).
Optimization of data collection taking radiation damage into account.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
409-419.
|
 |
|
|
|
|
 |
H.Y.Kaan,
V.Ulaganathan,
D.D.Hackney,
and
F.Kozielski
(2010).
An allosteric transition trapped in an intermediate state of a new kinesin-inhibitor complex.
|
| |
Biochem J,
425,
55-60.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.S.Paithankar,
and
E.F.Garman
(2010).
Know your dose: RADDOSE.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
381-388.
|
 |
|
|
|
|
 |
M.Schiltz,
and
G.Bricogne
(2010).
;Broken symmetries' in macromolecular crystallography: phasing from unmerged data.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
447-457.
|
 |
|
|
|
|
 |
M.Warkentin,
and
R.E.Thorne
(2010).
Glass transition in thaumatin crystals revealed through temperature-dependent radiation-sensitivity measurements.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
1092-1100.
|
 |
|
|
|
|
 |
P.Carpentier,
A.Royant,
M.Weik,
and
D.Bourgeois
(2010).
Raman-assisted crystallography suggests a mechanism of X-ray-induced disulfide radical formation and reparation.
|
| |
Structure,
18,
1410-1419.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Petrova,
S.Ginell,
A.Mitschler,
Y.Kim,
V.Y.Lunin,
G.Joachimiak,
A.Cousido-Siah,
I.Hazemann,
A.Podjarny,
K.Lazarski,
and
A.Joachimiak
(2010).
X-ray-induced deterioration of disulfide bridges at atomic resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
1075-1091.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.A.Kuntz,
W.Zhong,
J.Guo,
D.R.Rose,
and
G.J.Boons
(2009).
The Molecular Basis of Inhibition of Golgi alpha-Mannosidase II by Mannostatin A.
|
| |
Chembiochem,
10,
268-277.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.G.Wang,
P.K.Lee,
Y.H.Dong,
S.S.Pang,
R.G.Duggleby,
Z.M.Li,
and
L.W.Guddat
(2009).
Crystal structures of two novel sulfonylurea herbicides in complex with Arabidopsis thaliana acetohydroxyacid synthase.
|
| |
FEBS J,
276,
1282-1290.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Shi,
A.Proteau,
J.Wagner,
Q.Cui,
E.O.Purisima,
A.Matte,
and
M.Cygler
(2009).
Trapping open and closed forms of FitE: a group III periplasmic binding protein.
|
| |
Proteins,
75,
598-609.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Olieric,
U.Rieder,
K.Lang,
A.Serganov,
C.Schulze-Briese,
R.Micura,
P.Dumas,
and
E.Ennifar
(2009).
A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
|
| |
RNA,
15,
707-715.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Hong,
and
Q.Hao
(2009).
Measurements of accurate x-ray scattering data of protein solutions using small stationary sample cells.
|
| |
Rev Sci Instrum,
80,
014303.
|
 |
|
|
|
|
 |
D.S.Berkholz,
H.R.Faber,
S.N.Savvides,
and
P.A.Karplus
(2008).
Catalytic cycle of human glutathione reductase near 1 A resolution.
|
| |
J Mol Biol,
382,
371-384.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.P.Colletier,
D.Bourgeois,
B.Sanson,
D.Fournier,
J.L.Sussman,
I.Silman,
and
M.Weik
(2008).
Shoot-and-Trap: use of specific x-ray damage to study structural protein dynamics by temperature-controlled cryo-crystallography.
|
| |
Proc Natl Acad Sci U S A,
105,
11742-11747.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.M.Podust,
A.Ioanoviciu,
and
P.R.Ortiz de Montellano
(2008).
2.3 A X-ray structure of the heme-bound GAF domain of sensory histidine kinase DosT of Mycobacterium tuberculosis.
|
| |
Biochemistry,
47,
12523-12531.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.R.Roberts,
J.A.Tainer,
E.D.Getzoff,
D.A.Malencik,
S.R.Anderson,
V.C.Bomben,
K.R.Meyers,
P.A.Karplus,
and
J.S.Beckman
(2007).
Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS.
|
| |
J Mol Biol,
373,
877-890.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.J.Burden,
and
A.J.Oakley
(2007).
Anisotropic atomic motions in high-resolution protein crystallography molecular dynamics simulations.
|
| |
Phys Biol,
4,
79-90.
|
 |
|
|
|
|
 |
J.Gawronski-Salerno,
and
D.M.Freymann
(2007).
Structure of the GMPPNP-stabilized NG domain complex of the SRP GTPases Ffh and FtsY.
|
| |
J Struct Biol,
158,
122-128.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.R.Hobbs,
S.D.Munger,
and
G.L.Conn
(2007).
Monellin (MNEI) at 1.15 A resolution.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
162-167.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Standfuss,
G.Xie,
P.C.Edwards,
M.Burghammer,
D.D.Oprian,
and
G.F.Schertler
(2007).
Crystal structure of a thermally stable rhodopsin mutant.
|
| |
J Mol Biol,
372,
1179-1188.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.J.Southworth-Davies,
M.A.Medina,
I.Carmichael,
and
E.F.Garman
(2007).
Observation of decreased radiation damage at higher dose rates in room temperature protein crystallography.
|
| |
Structure,
15,
1531-1541.
|
 |
|
|
|
|
 |
H.Nakamichi,
and
T.Okada
(2006).
Local peptide movement in the photoreaction intermediate of rhodopsin.
|
| |
Proc Natl Acad Sci U S A,
103,
12729-12734.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Grabolle,
M.Haumann,
C.Müller,
P.Liebisch,
and
H.Dau
(2006).
Rapid loss of structural motifs in the manganese complex of oxygenic photosynthesis by X-ray irradiation at 10-300 K.
|
| |
J Biol Chem,
281,
4580-4588.
|
 |
|
|
|
|
 |
R.L.Owen,
E.Rudiño-Piñera,
and
E.F.Garman
(2006).
Experimental determination of the radiation dose limit for cryocooled protein crystals.
|
| |
Proc Natl Acad Sci U S A,
103,
4912-4917.
|
 |
|
|
|
|
 |
W.Blankenfeldt,
N.H.Thomä,
J.S.Wray,
M.Gautel,
and
I.Schlichting
(2006).
Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment.
|
| |
Proc Natl Acad Sci U S A,
103,
17713-17717.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.P.Dubnovitsky,
R.B.Ravelli,
A.N.Popov,
and
A.C.Papageorgiou
(2005).
Strain relief at the active site of phosphoserine aminotransferase induced by radiation damage.
|
| |
Protein Sci,
14,
1498-1507.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Volbeda,
L.Martin,
C.Cavazza,
M.Matho,
B.W.Faber,
W.Roseboom,
S.P.Albracht,
E.Garcin,
M.Rousset,
and
J.C.Fontecilla-Camps
(2005).
Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
|
| |
J Biol Inorg Chem,
10,
239-249.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Yano,
J.Kern,
K.D.Irrgang,
M.J.Latimer,
U.Bergmann,
P.Glatzel,
Y.Pushkar,
J.Biesiadka,
B.Loll,
K.Sauer,
J.Messinger,
A.Zouni,
and
V.K.Yachandra
(2005).
X-ray damage to the Mn4Ca complex in single crystals of photosystem II: a case study for metalloprotein crystallography.
|
| |
Proc Natl Acad Sci U S A,
102,
12047-12052.
|
 |
|
|
|
|
 |
M.E.Than,
G.P.Bourenkov,
S.Henrich,
K.Mann,
and
W.Bode
(2005).
The NC1 dimer of human placental basement membrane collagen IV: does a covalent crosslink exist?
|
| |
Biol Chem,
386,
759-766.
|
 |
|
|
|
|
 |
O.Carugo,
and
K.Djinović Carugo
(2005).
When X-rays modify the protein structure: radiation damage at work.
|
| |
Trends Biochem Sci,
30,
213-219.
|
 |
|
|
|
|
 |
R.M.Vanacore,
D.B.Friedman,
A.J.Ham,
M.Sundaramoorthy,
and
B.G.Hudson
(2005).
Identification of S-hydroxylysyl-methionine as the covalent cross-link of the noncollagenous (NC1) hexamer of the alpha1alpha1alpha2 collagen IV network: a role for the post-translational modification of lysine 211 to hydroxylysine 211 in hexamer assembly.
|
| |
J Biol Chem,
280,
29300-29310.
|
 |
|
|
|
|
 |
S.Covaceuszach,
A.Cattaneo,
and
D.Lamba
(2005).
Neutralization of NGF-TrkA receptor interaction by the novel antagonistic anti-TrkA monoclonal antibody MNAC13: a structural insight.
|
| |
Proteins,
58,
717-727.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Meunier-Jamin,
U.Kapp,
G.A.Leonard,
and
S.McSweeney
(2004).
The structure of the organic hydroperoxide resistance protein from Deinococcus radiodurans. Do conformational changes facilitate recycling of the redox disulfide?
|
| |
J Biol Chem,
279,
25830-25837.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Pechkova,
and
C.Nicolini
(2004).
Protein nanocrystallography: a new approach to structural proteomics.
|
| |
Trends Biotechnol,
22,
117-122.
|
 |
|
|
|
|
 |
R.Kort,
K.J.Hellingwerf,
and
R.B.Ravelli
(2004).
Initial events in the photocycle of photoactive yellow protein.
|
| |
J Biol Chem,
279,
26417-26424.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Anderson,
V.Srajer,
and
K.Moffat
(2004).
Structural heterogeneity of cryotrapped intermediates in the bacterial blue light photoreceptor, photoactive yellow protein.
|
| |
Photochem Photobiol,
80,
7.
|
 |
|
|
|
|
 |
M.S.Alphey,
M.Gabrielsen,
E.Micossi,
G.A.Leonard,
S.M.McSweeney,
R.B.Ravelli,
E.Tetaud,
A.H.Fairlamb,
C.S.Bond,
and
W.N.Hunter
(2003).
Tryparedoxins from Crithidia fasciculata and Trypanosoma brucei: photoreduction of the redox disulfide using synchrotron radiation and evidence for a conformational switch implicated in function.
|
| |
J Biol Chem,
278,
25919-25925.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Sliz,
S.C.Harrison,
and
G.Rosenbaum
(2003).
How does radiation damage in protein crystals depend on X-ray dose?
|
| |
Structure,
11,
13-19.
|
 |
|
|
|
|
 |
P.T.Erskine,
L.Coates,
S.Mall,
R.S.Gill,
S.P.Wood,
D.A.Myles,
and
J.B.Cooper
(2003).
Atomic resolution analysis of the catalytic site of an aspartic proteinase and an unexpected mode of binding by short peptides.
|
| |
Protein Sci,
12,
1741-1749.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Fedorov,
I.Schlichting,
E.Hartmann,
T.Domratcheva,
M.Fuhrmann,
and
P.Hegemann
(2003).
Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
|
| |
Biophys J,
84,
2474-2482.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Della Longa,
A.Arcovito,
M.Benfatto,
A.Congiu-Castellano,
M.Girasole,
J.L.Hazemann,
and
A.Lo Bosco
(2003).
Redox-induced structural dynamics of Fe-heme ligand in myoglobin by X-ray absorption spectroscopy.
|
| |
Biophys J,
85,
549-558.
|
 |
|
|
|
|
 |
S.Parthasarathy,
K.Eaazhisai,
H.Balaram,
P.Balaram,
and
M.R.Murthy
(2003).
Structure of Plasmodium falciparum triose-phosphate isomerase-2-phosphoglycerate complex at 1.1-A resolution.
|
| |
J Biol Chem,
278,
52461-52470.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.S.Pang,
L.W.Guddat,
and
R.G.Duggleby
(2003).
Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase.
|
| |
J Biol Chem,
278,
7639-7644.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.M.Wilmot,
and
A.R.Pearson
(2002).
Cryocrystallography of metalloprotein reaction intermediates.
|
| |
Curr Opin Chem Biol,
6,
202-207.
|
 |
|
|
|
|
 |
J.J.van Thor,
T.Gensch,
K.J.Hellingwerf,
and
L.N.Johnson
(2002).
Phototransformation of green fluorescent protein with UV and visible light leads to decarboxylation of glutamate 222.
|
| |
Nat Struct Biol,
9,
37-41.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.V.Hartemann,
H.A.Baldis,
A.K.Kerman,
A.Le Foll,
N.C.Luhmann,
and
B.Rupp
(2001).
Three-dimensional theory of emittance in Compton scattering and x-ray protein crystallography.
|
| |
Phys Rev E Stat Nonlin Soft Matter Phys,
64,
016501.
|
 |
|
|
|
|
 |
M.Weik,
R.B.Ravelli,
I.Silman,
J.L.Sussman,
P.Gros,
and
J.Kroon
(2001).
Specific protein dynamics near the solvent glass transition assayed by radiation-induced structural changes.
|
| |
Protein Sci,
10,
1953-1961.
|
 |
|
|
|
|
 |
P.J.Ellis,
T.Conrads,
R.Hille,
and
P.Kuhn
(2001).
Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
|
| |
Structure,
9,
125-132.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Gaboriaud,
V.Rossi,
I.Bally,
G.J.Arlaud,
and
J.C.Fontecilla-Camps
(2000).
Crystal structure of the catalytic domain of human complement c1s: a serine protease with a handle.
|
| |
EMBO J,
19,
1755-1765.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

