 |
PDBsum entry 3djg

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3djg
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.1.7
- glutathione-disulfide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 glutathione + NADP+ = glutathione disulfide + NADPH + H+
|
 |
 |
 |
 |
 |
 2
×
glutathione
2
×
glutathione
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
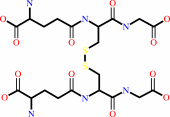 glutathione disulfide
glutathione disulfide
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
382:371-384
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Catalytic cycle of human glutathione reductase near 1 A resolution.
|
|
D.S.Berkholz,
H.R.Faber,
S.N.Savvides,
P.A.Karplus.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Efficient enzyme catalysis depends on exquisite details of structure beyond
those resolvable in typical medium- and high-resolution crystallographic
analyses. Here we report synchrotron-based cryocrystallographic studies of
natural substrate complexes of the flavoenzyme human glutathione reductase (GR)
at nominal resolutions between 1.1 and 0.95 A that reveal new aspects of its
mechanism. Compression in the active site causes overlapping van der Waals radii
and distortion in the nicotinamide ring of the NADPH substrate, which enhances
catalysis via stereoelectronic effects. The bound NADPH and redox-active
disulfide are positioned optimally on opposite sides of the flavin for a
1,2-addition across a flavin double bond. The new structures extend earlier
observations to reveal that the redox-active disulfide loop in GR is an extreme
case of sequential peptide bonds systematically deviating from planarity--a net
deviation of 53 degrees across five residues. But this apparent strain is not a
factor in catalysis, as it is present in both oxidized and reduced structures.
Intriguingly, the flavin bond lengths in oxidized GR are intermediate between
those expected for oxidized and reduced flavin, but we present evidence that
this may not be due to the protein environment but instead due to partial
synchrotron reduction of the flavin by the synchrotron beam. Finally, of more
general relevance, we present evidence that the structures of
synchrotron-reduced disulfide bonds cannot generally be used as reliable models
for naturally reduced disulfide bonds.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 7.
Fig. 7. Nicotinamide distortion and ribose conformation favor
catalysis. (a) The schematic shows the planes of the
nicotinamide and flavin (solid black lines). The hypothesized
partial boat is shown as a solid red line. Pyramidalization at
the nicotinamide N1 places the lone pair on the flavin side,
where it (i) entropically favors the productive boat
conformation to form, and (ii) repels the hydride to be
transferred (dashed red line). (b) The ribose conformation
relative to the nicotinamide stabilizes the electron-deficient
NADP^+ ring orbitals via hyperconjugative electron donation from
the ribose. The glycosidic C–O bond position parallel with the
nicotinamide ring also favors NADP^+ over NADPH (see Results and
Discussion).
|
 |
Figure 8.
Fig. 8. Stereoelectronic control in nicotinamide–flavin
interaction. (a) A side view with the flavin N5–C4a bond in
the plane of the paper and (b) a view down the flavin N5–C4a
bond together show the optimal geometry for concerted
1,2-addition across the double bond. Compression in the form of
shorter-than-van-der-Waals interactions is also shown in (a).
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Elsevier:
J Mol Biol
(2008,
382,
371-384)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.Vergauwen,
J.Elegheert,
A.Dansercoer,
B.Devreese,
and
S.N.Savvides
(2010).
Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA.
|
| |
Proc Natl Acad Sci U S A,
107,
13270-13275.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.S.Berkholz,
P.B.Krenesky,
J.R.Davidson,
and
P.A.Karplus
(2010).
Protein Geometry Database: a flexible engine to explore backbone conformations and their relationships to covalent geometry.
|
| |
Nucleic Acids Res,
38,
D320-D325.
|
 |
|
|
|
|
 |
N.Satoh,
N.Watanabe,
A.Kanda,
M.Sugaya-Fukasawa,
and
H.Hisatomi
(2010).
Expression of glutathione reductase splice variants in human tissues.
|
| |
Biochem Genet,
48,
816-821.
|
 |
|
|
|
|
 |
D.S.Berkholz,
M.V.Shapovalov,
R.L.Dunbrack,
and
P.A.Karplus
(2009).
Conformation dependence of backbone geometry in proteins.
|
| |
Structure,
17,
1316-1325.
|
 |
|
|
|
|
 |
J.R.Wallen,
T.C.Mallett,
W.Boles,
D.Parsonage,
C.M.Furdui,
P.A.Karplus,
and
A.Claiborne
(2009).
Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
|
| |
Biochemistry,
48,
9650-9667.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.A.Kashem,
H.D.Etages,
N.Kopitar-Jerala,
I.S.McGregor,
and
I.Matsumoto
(2009).
Differential protein expression in the corpus callosum (body) of human alcoholic brain.
|
| |
J Neurochem,
110,
486-495.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

