 |
PDBsum entry 1aln

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.5
- cytidine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
cytidine + H2O + H+ = uridine + NH4+
|
|
2.
|
2'-deoxycytidine + H2O + H+ = 2'-deoxyuridine + NH4+
|
|
 |
 |
 |
 |
 |
cytidine
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
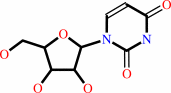 uridine
uridine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
2'-deoxycytidine
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
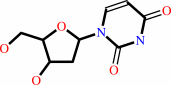 2'-deoxyuridine
2'-deoxyuridine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
35:1335-1341
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Cytidine deaminase complexed to 3-deazacytidine: a "valence buffer" in zinc enzyme catalysis.
|
|
S.Xiang,
S.A.Short,
R.Wolfenden,
C.W.Carter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The cytidine deaminase substrate analog inhibitor 3-deazacytidine binds with its
4-amino group inserted into a site previously identified as a probable binding
site for the leaving ammonia group. Binding to this site shifts the pyrimidine
ring significantly further from the activated water molecule than the position
it occupies in either of two complexes with compounds capable of hydrogen
bonding at the 3-position of the ring [Xiang et al. (1995) Biochemistry 34,
4516-4523]. Difference Fourier maps between the deazacytidine,
dihydrozebularine, and zebularine--hydrate inhibitor complexes suggest that the
ring itself moves successively toward the activated water, leaving the amino
group behind in this site as the substrate complex approaches the transition
state. They also reveal systematic changes in a single zinc-sulfur bond
distance. These correlate with chemical changes expected as the substrate
approaches the tetrahedral transition state, in which the zinc-activated
hydroxyl group develops maximal negative charge and forms a short hydrogen bond
to the neighboring carboxylate group of Glu 104. Empirical bond valence
relationships suggest that the Zn-S gamma 132 bond functions throughout the
reaction as a "valence buffer" that accommodates changing negative charge on the
hydroxyl group. Similar structural features in alcohol dehydrogenase suggest
that analogous mechanisms may be a general feature of catalysis by zinc enzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
X.Li,
S.A.Hayik,
and
K.M.Merz
(2010).
QM/MM X-ray refinement of zinc metalloenzymes.
|
| |
J Inorg Biochem,
104,
512-522.
|
 |
|
|
|
|
 |
M.Koutmos,
R.Pejchal,
T.M.Bomer,
R.G.Matthews,
J.L.Smith,
and
M.L.Ludwig
(2008).
Metal active site elasticity linked to activation of homocysteine in methionine synthases.
|
| |
Proc Natl Acad Sci U S A,
105,
3286-3291.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Kumasaka,
M.Yamamoto,
M.Furuichi,
M.Nakasako,
A.H.Teh,
M.Kimura,
I.Yamaguchi,
and
T.Ueki
(2007).
Crystal Structures of Blasticidin S Deaminase (BSD): IMPLICATIONS FOR DYNAMIC PROPERTIES OF CATALYTIC ZINC.
|
| |
J Biol Chem,
282,
37103-37111.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Schwartz,
and
J.King
(2006).
Frequencies of hydrophobic and hydrophilic runs and alternations in proteins of known structure.
|
| |
Protein Sci,
15,
102-112.
|
 |
|
|
|
|
 |
A.J.Bordner,
and
R.Abagyan
(2005).
Statistical analysis and prediction of protein-protein interfaces.
|
| |
Proteins,
60,
353-366.
|
 |
|
|
|
|
 |
J.E.Tomassini,
K.Getty,
M.W.Stahlhut,
S.Shim,
B.Bhat,
A.B.Eldrup,
T.P.Prakash,
S.S.Carroll,
O.Flores,
M.MacCoss,
D.R.McMasters,
G.Migliaccio,
and
D.B.Olsen
(2005).
Inhibitory effect of 2'-substituted nucleosides on hepatitis C virus replication correlates with metabolic properties in replicon cells.
|
| |
Antimicrob Agents Chemother,
49,
2050-2058.
|
 |
|
|
|
|
 |
R.Pejchal,
and
M.L.Ludwig
(2005).
Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication.
|
| |
PLoS Biol,
3,
e31.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Qi,
and
N.V.Grishin
(2005).
Structural classification of thioredoxin-like fold proteins.
|
| |
Proteins,
58,
376-388.
|
 |
|
|
|
|
 |
K.Xie,
M.P.Sowden,
G.S.Dance,
A.T.Torelli,
H.C.Smith,
and
J.E.Wedekind
(2004).
The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1.
|
| |
Proc Natl Acad Sci U S A,
101,
8114-8119.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Kojima,
K.Inoue,
R.Nakajima-Shibata,
S.Kawahara,
and
E.Ohtsuka
(2003).
A new, but old, nucleoside analog: the first synthesis of 1-deaza-2'-deoxyguanosine and its properties as a nucleoside and as oligodeoxynucleotides.
|
| |
Nucleic Acids Res,
31,
7175-7188.
|
 |
|
|
|
|
 |
T.P.Ko,
J.J.Lin,
C.Y.Hu,
Y.H.Hsu,
A.H.Wang,
and
S.H.Liaw
(2003).
Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution.
|
| |
J Biol Chem,
278,
19111-19117.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Snider,
D.Lazarevic,
and
R.Wolfenden
(2002).
Catalysis by entropic effects: the action of cytidine deaminase on 5,6-dihydrocytidine.
|
| |
Biochemistry,
41,
3925-3930.
|
 |
|
|
|
|
 |
R.C.Noonan,
C.W.Carter CW,
and
C.K.Bagdassarian
(2002).
Enzymatic conformational fluctuations along the reaction coordinate of cytidine deaminase.
|
| |
Protein Sci,
11,
1424-1434.
|
 |
|
|
|
|
 |
K.O.Alper,
M.Singla,
J.L.Stone,
and
C.K.Bagdassarian
(2001).
Correlated conformational fluctuations during enzymatic catalysis: Implications for catalytic rate enhancement.
|
| |
Protein Sci,
10,
1319-1330.
|
 |
|
|
|
|
 |
A.Somasekaram,
A.Jarmuz,
A.How,
J.Scott,
and
N.Navaratnam
(1999).
Intracellular localization of human cytidine deaminase. Identification of a functional nuclear localization signal.
|
| |
J Biol Chem,
274,
28405-28412.
|
 |
|
|
|
|
 |
D.C.Carlow,
S.A.Short,
and
R.Wolfenden
(1998).
Complementary truncations of a hydrogen bond to ribose involved in transition-state stabilization by cytidine deaminase.
|
| |
Biochemistry,
37,
1199-1203.
|
 |
|
|
|
|
 |
Z.Wang,
and
F.A.Quiocho
(1998).
Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity.
|
| |
Biochemistry,
37,
8314-8324.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.W.Adolph,
M.Kiefer,
and
E.Cedergren-Zeppezauer
(1997).
Electrostatic effects in the kinetics of coenzyme binding to isozymes of alcohol dehydrogenase from horse liver.
|
| |
Biochemistry,
36,
8743-8754.
|
 |
|
|
|
|
 |
W.Huang,
J.Jia,
J.Cummings,
M.Nelson,
G.Schneider,
and
Y.Lindqvist
(1997).
Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold.
|
| |
Structure,
5,
691-699.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Grosjean,
S.Auxilien,
F.Constantinesco,
C.Simon,
Y.Corda,
H.F.Becker,
D.Foiret,
A.Morin,
Y.X.Jin,
M.Fournier,
and
J.L.Fourrey
(1996).
Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review.
|
| |
Biochimie,
78,
488-501.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

