 |
PDBsum entry 1a4m

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.4
- adenosine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
adenosine + H2O + H+ = inosine + NH4+
|
|
2.
|
2'-deoxyadenosine + H2O + H+ = 2'-deoxyinosine + NH4+
|
|
 |
 |
 |
 |
 |
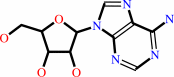 adenosine
adenosine
|
+
|
H2O
|
+
|
H(+)
|
=
|
inosine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
2'-deoxyadenosine
Bound ligand (Het Group name = )
matches with 85.00% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyinosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:8314-8324
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity.
|
|
Z.Wang,
F.A.Quiocho.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Adenosine deaminase, which catalyzes the irreversible hydrolytic deamination of
adenosine nucleosides to inosine nucleosides and ammonia, is a key enzyme in
purine metabolism and lymphoid development. The X-ray structures of murine
adenosine deaminase with bound potent inhibitors (Ki values approximately
10(-13) M) (8R)-hydroxyl-2'-deoxycoformycin (pentostatin), a transition state
analogue, and (6S)-hydroxyl-1,6-dihydropurine riboside, a reaction coordinate
analogue, have been determined and refined to resolutions of 2.6 and 1.95 A,
respectively. Crystals of both complexes were obtained at pH 7, where the enzyme
is fully active, in an identical space group with the asymmetric unit containing
four molecules. In addition to the very high degree of similarity between the
four independent molecules in each complex structure, there is also considerable
structural similarity of the complex with the dihydropurine riboside with that
of an identical complex previously determined at pH 4.2 where the enzyme is 20%
active. The interactions between the enzyme and the two analogues are extremely
similar. These include the coordination of the 8R- or 6S-hydroxyl group of the
analogues to the Zn2+ which mainly contributes to the strong potency and very
high degree of stereospecificity of inhibition by these analogues. The
interactions are further indicative of the structural and chemical requirements
of substrates. These structures and recent site-directed mutagenesis have
further shed light on the catalytic mechanism of the enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.F.Xiang,
Y.Patskovsky,
C.Xu,
A.J.Meyer,
J.M.Sauder,
S.K.Burley,
S.C.Almo,
and
F.M.Raushel
(2009).
Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
|
| |
Biochemistry,
48,
3730-3742.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Y.Little,
and
L.Chen
(2009).
Identification of coevolving residues and coevolution potentials emphasizing structure, bond formation and catalytic coordination in protein evolution.
|
| |
PLoS ONE,
4,
e4762.
|
 |
|
|
|
|
 |
M.C.Ho,
M.B.Cassera,
D.C.Madrid,
L.M.Ting,
P.C.Tyler,
K.Kim,
S.C.Almo,
and
V.L.Schramm
(2009).
Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases.
|
| |
Biochemistry,
48,
9618-9626.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.R.Ludek,
G.K.Schroeder,
C.Liao,
P.L.Russ,
R.Wolfenden,
and
V.E.Marquez
(2009).
Synthesis and conformational analysis of locked carbocyclic analogues of 1,3-diazepinone riboside, a high-affinity cytidine deaminase inhibitor.
|
| |
J Org Chem,
74,
6212-6223.
|
 |
|
|
|
|
 |
V.E.Marquez,
G.K.Schroeder,
O.R.Ludek,
M.A.Siddiqui,
A.Ezzitouni,
and
R.Wolfenden
(2009).
Contrasting behavior of conformationally locked carbocyclic nucleosides of adenosine and cytidine as substrates for deaminases.
|
| |
Nucleosides Nucleotides Nucleic Acids,
28,
614-632.
|
 |
|
|
|
|
 |
E.T.Larson,
W.Deng,
B.E.Krumm,
A.Napuli,
N.Mueller,
W.C.Van Voorhis,
F.S.Buckner,
E.Fan,
A.Lauricella,
G.DeTitta,
J.Luft,
F.Zucker,
W.G.Hol,
C.L.Verlinde,
and
E.A.Merritt
(2008).
Structures of substrate- and inhibitor-bound adenosine deaminase from a human malaria parasite show a dramatic conformational change and shed light on drug selectivity.
|
| |
J Mol Biol,
381,
975-988.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Butora,
D.B.Olsen,
S.S.Carroll,
D.R.McMasters,
C.Schmitt,
J.F.Leone,
M.Stahlhut,
C.Burlein,
and
M.Maccoss
(2007).
Synthesis and HCV inhibitory properties of 9-deaza- and 7,9-dideaza-7-oxa-2'-C-methyladenosine.
|
| |
Bioorg Med Chem,
15,
5219-5229.
|
 |
|
|
|
|
 |
S.Escusa,
D.Laporte,
A.Massoni,
H.Boucherie,
A.Dautant,
and
B.Daignan-Fornier
(2007).
Skp1-Cullin-F-box-dependent degradation of Aah1p requires its interaction with the F-box protein Saf1p.
|
| |
J Biol Chem,
282,
20097-20103.
|
 |
|
|
|
|
 |
B.W.Han,
C.A.Bingman,
D.K.Mahnke,
R.M.Bannen,
S.Y.Bednarek,
R.L.Sabina,
and
G.N.Phillips
(2006).
Membrane association, mechanism of action, and structure of Arabidopsis embryonic factor 1 (FAC1).
|
| |
J Biol Chem,
281,
14939-14947.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Goto,
H.Hayashi,
I.Miyahara,
K.Hirotsu,
M.Yoshida,
and
T.Oikawa
(2006).
Crystal structures of nonoxidative zinc-dependent 2,6-dihydroxybenzoate (gamma-resorcylate) decarboxylase from Rhizobium sp. strain MTP-10005.
|
| |
J Biol Chem,
281,
34365-34373.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.A.Il'icheva,
I.u.P.Zarubin,
P.A.Kostin,
D.V.Mirgorodskiĭ,
P.P.Purygin,
and
V.L.Florent'ev
(2005).
[Theoretical study of the structure of adenosine deaminase complexes with adenosine analogues: I. Aza-, deaza- and isomeric azadeazaanalogues of adenosine]
|
| |
Bioorg Khim,
31,
488-502.
|
 |
|
|
|
|
 |
K.Wang,
and
R.Samudrala
(2005).
FSSA: a novel method for identifying functional signatures from structural alignments.
|
| |
Bioinformatics,
21,
2969-2977.
|
 |
|
|
|
|
 |
S.A.Maier,
J.R.Galellis,
and
H.E.McDermid
(2005).
Phylogenetic analysis reveals a novel protein family closely related to adenosine deaminase.
|
| |
J Mol Evol,
61,
776-794.
|
 |
|
|
|
|
 |
F.Vincent,
D.Yates,
E.Garman,
G.J.Davies,
and
J.A.Brannigan
(2004).
The three-dimensional structure of the N-acetylglucosamine-6-phosphate deacetylase, NagA, from Bacillus subtilis: a member of the urease superfamily.
|
| |
J Biol Chem,
279,
2809-2816.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.C.Deo,
E.F.Schmidt,
A.Elhabazi,
H.Togashi,
S.K.Burley,
and
S.M.Strittmatter
(2004).
Structural bases for CRMP function in plexin-dependent semaphorin3A signaling.
|
| |
EMBO J,
23,
9.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.P.Ko,
J.J.Lin,
C.Y.Hu,
Y.H.Hsu,
A.H.Wang,
and
S.H.Liaw
(2003).
Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution.
|
| |
J Biol Chem,
278,
19111-19117.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Cristalli,
S.Costanzi,
C.Lambertucci,
G.Lupidi,
S.Vittori,
R.Volpini,
and
E.Camaioni
(2001).
Adenosine deaminase: functional implications and different classes of inhibitors.
|
| |
Med Res Rev,
21,
105-128.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
D.K.Leung,
Z.Yang,
and
R.Breslow
(2000).
Selective disruption of protein aggregation by cyclodextrin dimers.
|
| |
Proc Natl Acad Sci U S A,
97,
5050-5053.
|
 |
|
|
|
|
 |
H.Ford,
F.Dai,
L.Mu,
M.A.Siddiqui,
M.C.Nicklaus,
L.Anderson,
V.E.Marquez,
and
J.J.Barchi
(2000).
Adenosine deaminase prefers a distinct sugar ring conformation for binding and catalysis: kinetic and structural studies.
|
| |
Biochemistry,
39,
2581-2592.
|
 |
|
|
|
|
 |
U.Ryde
(1999).
Carboxylate binding modes in zinc proteins: A theoretical study
|
| |
Biophys J,
77,
2777-2787.
|
 |
|
|
|
|
 |
U.Ermler,
W.Grabarse,
S.Shima,
M.Goubeaud,
and
R.K.Thauer
(1998).
Active sites of transition-metal enzymes with a focus on nickel.
|
| |
Curr Opin Struct Biol,
8,
749-758.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

