 |
PDBsum entry 1uaq

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.1
- cytosine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cytosine + H2O + H+ = uracil + NH4+
|
 |
 |
 |
 |
 |
 cytosine
cytosine
|
+
|
H2O
|
+
|
H(+)
|
=
|
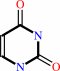 uracil
uracil
|
+
|
NH4(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:19111-19117
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution.
|
|
T.P.Ko,
J.J.Lin,
C.Y.Hu,
Y.H.Hsu,
A.H.Wang,
S.H.Liaw.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Yeast cytosine deaminase is an attractive candidate for anticancer gene therapy
because it catalyzes the deamination of the prodrug 5-fluorocytosine to form
5-fluorouracil. We report here the crystal structure of the enzyme in complex
with the inhibitor 2-hydroxypyrimidine at 1.6-A resolution. The protein forms a
tightly packed dimer with an extensive interface of 1450 A2 per monomer. The
inhibitor was converted into a hydrated adduct as a transition-state analog. The
essential zinc ion is ligated by the 4-hydroxyl group of the inhibitor together
with His62, Cys91, and Cys94 from the protein. The enzyme shares similar
active-site architecture to cytidine deaminases and an unusually high structural
homology to 5-aminoimidazole-4-carboxamide-ribonucleotide transformylase and
thereby may define a new superfamily. The unique C-terminal tail is involved in
substrate specificity and also functions as a gate controlling access to the
active site. The complex structure reveals a closed conformation, suggesting
that substrate binding seals the active-site entrance so that the catalytic
groups are sequestered from solvent. A comparison of the crystal structures of
the bacterial and fungal cytosine deaminases provides an elegant example of
convergent evolution, where starting from unrelated ancestral proteins, the same
metal-assisted deamination is achieved through opposite chiral intermediates
within distinctly different active sites.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. Structure of yeast CD. A, stereo view of the
monomer, which is a three-layered  / /  / /  structure with a
central structure with a
central  -sheet sandwiched on
either side by -sheet sandwiched on
either side by  -helices. The tightly
bound zinc ion is shown as a magenta sphere with its ligands and
the inhibitor (3,4-dihydrouracil (DHU)) as ball-and-stick
representations. B, the dimer has one monomer colored in red
(helices) and green (strands), whereas the other is colored in
magenta (helices) and blue (strands). The zinc ions are shown as
yellow spheres. -helices. The tightly
bound zinc ion is shown as a magenta sphere with its ligands and
the inhibitor (3,4-dihydrouracil (DHU)) as ball-and-stick
representations. B, the dimer has one monomer colored in red
(helices) and green (strands), whereas the other is colored in
magenta (helices) and blue (strands). The zinc ions are shown as
yellow spheres.
|
 |
Figure 3.
FIG. 3. The active site. A, stereo view of the 2F[o] - F[c]
electron density map of the active site contoured at 1.5  level
and shown in cyan and the difference anomalous map for the zinc
ion contoured at 30 level
and shown in cyan and the difference anomalous map for the zinc
ion contoured at 30  level and shown in
purple. The densities of the inhibitor molecule are highlighted
in green. The active-site residues and the inhibitor
(3,4-dihydrouracil (DHU)) are shown as ball-and-stick
representations, and the zinc ion is shown as a magenta sphere.
B, stereo view of the interaction networks in the active site.
There are six direct hydrogen bonds between the protein molecule
and the inhibitor (see The Active Site Architecture and
Substrate Binding). C, stereo view of a comparison of the active
sites of the yeast (magenta) and E. coli cytosine deaminase
(Protein Data Bank code 1K70 [PDB]
, green) based on superposition of the inhibitor
3,4-dihydrouracil. The residue numbering is labeled in the same
color for each protein. The bacterial enzymes utilize three
histidines and one aspartate for iron ligation (His61, His63,
His214, and Asp313). Asp313 also forms a hydrogen bond with the
attacking water molecule. Glu217 interacts with the N3 atom of
the pyrimidine ring, and Gln156 interacts with the O2 and N1
atoms. level and shown in
purple. The densities of the inhibitor molecule are highlighted
in green. The active-site residues and the inhibitor
(3,4-dihydrouracil (DHU)) are shown as ball-and-stick
representations, and the zinc ion is shown as a magenta sphere.
B, stereo view of the interaction networks in the active site.
There are six direct hydrogen bonds between the protein molecule
and the inhibitor (see The Active Site Architecture and
Substrate Binding). C, stereo view of a comparison of the active
sites of the yeast (magenta) and E. coli cytosine deaminase
(Protein Data Bank code 1K70 [PDB]
, green) based on superposition of the inhibitor
3,4-dihydrouracil. The residue numbering is labeled in the same
color for each protein. The bacterial enzymes utilize three
histidines and one aspartate for iron ligation (His61, His63,
His214, and Asp313). Asp313 also forms a hydrogen bond with the
attacking water molecule. Glu217 interacts with the N3 atom of
the pyrimidine ring, and Gln156 interacts with the O2 and N1
atoms.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
19111-19117)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.E.Metzger,
J.K.Lee,
J.S.Finer-Moore,
C.R.Raetz,
and
R.M.Stroud
(2012).
LpxI structures reveal how a lipid A precursor is synthesized.
|
| |
Nat Struct Mol Biol,
19,
1132-1138.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Vandeputte,
L.Pineau,
G.Larcher,
T.Noel,
D.Brèthes,
D.Chabasse,
and
J.P.Bouchara
(2011).
Molecular mechanisms of resistance to 5-fluorocytosine in laboratory mutants of Candida glabrata.
|
| |
Mycopathologia,
171,
11-21.
|
 |
|
|
|
|
 |
B.A.McManus,
G.P.Moran,
J.A.Higgins,
D.J.Sullivan,
and
D.C.Coleman
(2009).
A Ser29Leu substitution in the cytosine deaminase Fca1p is responsible for clade-specific flucytosine resistance in Candida dubliniensis.
|
| |
Antimicrob Agents Chemother,
53,
4678-4685.
|
 |
|
|
|
|
 |
J.I.Park,
L.Cao,
V.M.Platt,
Z.Huang,
R.A.Stull,
E.E.Dy,
J.J.Sperinde,
J.S.Yokoyama,
and
F.C.Szoka
(2009).
Antitumor therapy mediated by 5-fluorocytosine and a recombinant fusion protein containing TSG-6 hyaluronan binding domain and yeast cytosine deaminase.
|
| |
Mol Pharm,
6,
801-812.
|
 |
|
|
|
|
 |
M.Florent,
T.Noël,
G.Ruprich-Robert,
B.Da Silva,
V.Fitton-Ouhabi,
C.Chastin,
N.Papon,
and
F.Chapeland-Leclerc
(2009).
Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae.
|
| |
Antimicrob Agents Chemother,
53,
2982-2990.
|
 |
|
|
|
|
 |
M.Henry,
D.Guétard,
R.Suspène,
C.Rusniok,
S.Wain-Hobson,
and
J.P.Vartanian
(2009).
Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G.
|
| |
PLoS ONE,
4,
e4277.
|
 |
|
|
|
|
 |
A.Liu,
J.Wang,
Z.Lu,
L.Yao,
Y.Li,
and
H.Yan
(2008).
Hydrogen-bond detection, configuration assignment and rotamer correction of side-chain amides in large proteins by NMR spectroscopy through protium/deuterium isotope effects.
|
| |
Chembiochem,
9,
2860-2871.
|
 |
|
|
|
|
 |
D.Wolf,
and
S.P.Goff
(2008).
Host restriction factors blocking retroviral replication.
|
| |
Annu Rev Genet,
42,
143-163.
|
 |
|
|
|
|
 |
K.M.Chen,
E.Harjes,
P.J.Gross,
A.Fahmy,
Y.Lu,
K.Shindo,
R.S.Harris,
and
H.Matsuo
(2008).
Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G.
|
| |
Nature,
452,
116-119.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.S.Brar,
E.J.Sacho,
I.Tessmer,
D.L.Croteau,
D.A.Erie,
and
M.Diaz
(2008).
Activation-induced deaminase, AID, is catalytically active as a monomer on single-stranded DNA.
|
| |
DNA Repair (Amst),
7,
77-87.
|
 |
|
|
|
|
 |
T.Matsubara,
M.Dupuis,
and
M.Aida
(2008).
An insight into the environmental effects of the pocket of the active site of the enzyme. Ab initio ONIOM-molecular dynamics (MD) study on cytosine deaminase.
|
| |
J Comput Chem,
29,
458-465.
|
 |
|
|
|
|
 |
E.Johansson,
M.Thymark,
J.H.Bynck,
M.Fanø,
S.Larsen,
and
M.Willemoës
(2007).
Regulation of dCTP deaminase from Escherichia coli by nonallosteric dTTP binding to an inactive form of the enzyme.
|
| |
FEBS J,
274,
4188-4198.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Huthoff,
and
M.H.Malim
(2007).
Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation.
|
| |
J Virol,
81,
3807-3815.
|
 |
|
|
|
|
 |
J.Dechancie,
F.R.Clemente,
A.J.Smith,
H.Gunaydin,
Y.L.Zhao,
X.Zhang,
and
K.N.Houk
(2007).
How similar are enzyme active site geometries derived from quantum mechanical theozymes to crystal structures of enzyme-inhibitor complexes? Implications for enzyme design.
|
| |
Protein Sci,
16,
1851-1866.
|
 |
|
|
|
|
 |
L.Yao,
H.Yan,
and
R.I.Cukier
(2007).
A molecular dynamics study of the ligand release path in yeast cytosine deaminase.
|
| |
Biophys J,
92,
2301-2310.
|
 |
|
|
|
|
 |
A.Liu,
Y.Li,
L.Yao,
and
H.Yan
(2006).
Simultaneous NMR assignment of backbone and side chain amides in large proteins with IS-TROSY.
|
| |
J Biomol NMR,
36,
205-214.
|
 |
|
|
|
|
 |
M.Kato,
R.M.Wynn,
J.L.Chuang,
C.A.Brautigam,
M.Custorio,
and
D.T.Chuang
(2006).
A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
|
| |
EMBO J,
25,
5983-5994.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Kim,
N.Maltseva,
I.Dementieva,
F.Collart,
D.Holzle,
and
A.Joachimiak
(2006).
Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 A resolution.
|
| |
Proteins,
63,
1097-1101.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.P.Xiong,
and
G.S.Kwon
(2005).
PEGylation of yeast cytosine deaminase for pretargeting.
|
| |
J Pharm Sci,
94,
1249-1258.
|
 |
|
|
|
|
 |
Q.Xu,
R.Schwarzenbacher,
D.McMullan,
P.Abdubek,
E.Ambing,
T.Biorac,
J.M.Canaves,
H.J.Chiu,
X.Dai,
A.M.Deacon,
M.DiDonato,
M.A.Elsliger,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
E.Hampton,
M.Hornsby,
L.Jaroszewski,
H.E.Klock,
E.Koesema,
A.Kreusch,
P.Kuhn,
S.A.Lesley,
I.Levin,
M.D.Miller,
A.Morse,
K.Moy,
J.Ouyang,
R.Page,
K.Quijano,
R.Reyes,
A.Robb,
E.Sims,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
F.von Delft,
X.Wang,
B.West,
A.White,
G.Wolf,
O.Zagnitko,
K.O.Hodgson,
J.Wooley,
and
I.A.Wilson
(2005).
Crystal structure of a formiminotetrahydrofolate cyclodeaminase (TM1560) from Thermotoga maritima at 2.80 A resolution reveals a new fold.
|
| |
Proteins,
58,
976-981.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Shen,
N.L.Zhukovskaya,
Q.Guo,
J.Florián,
and
W.J.Tang
(2005).
Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
|
| |
EMBO J,
24,
929-941.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Xie,
M.P.Sowden,
G.S.Dance,
A.T.Torelli,
H.C.Smith,
and
J.E.Wedekind
(2004).
The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1.
|
| |
Proc Natl Acad Sci U S A,
101,
8114-8119.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.S.Harris,
and
M.T.Liddament
(2004).
Retroviral restriction by APOBEC proteins.
|
| |
Nat Rev Immunol,
4,
868-877.
|
 |
|
|
|
|
 |
Y.J.Chang,
C.H.Huang,
C.Y.Hu,
and
S.H.Liaw
(2004).
Crystallization and preliminary crystallographic analysis of Bacillus subtilis guanine deaminase.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1152-1154.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

