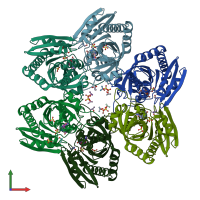
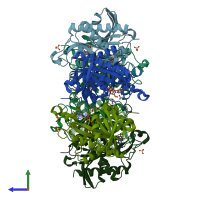
Assemblies
Assembly Name:
Purine nucleoside phosphorylase
Multimeric state:
homo hexamer
Accessible surface area:
45437.41 Å2
Buried surface area:
32701.62 Å2
Dissociation area:
8,082.08
Å2
Dissociation energy (ΔGdiss):
28.7
kcal/mol
Dissociation entropy (TΔSdiss):
30.37
kcal/mol
Symmetry number:
6
PDBe Complex ID:
PDB-CPX-184666
Assembly Name:
Purine nucleoside phosphorylase
Multimeric state:
homo dimer
Accessible surface area:
19935.51 Å2
Buried surface area:
5864.2 Å2
Dissociation area:
1,342.22
Å2
Dissociation energy (ΔGdiss):
16.44
kcal/mol
Dissociation entropy (TΔSdiss):
13.09
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-184665
Assembly Name:
Purine nucleoside phosphorylase
Multimeric state:
homo dimer
Accessible surface area:
19878.45 Å2
Buried surface area:
6639.7 Å2
Dissociation area:
1,338.52
Å2
Dissociation energy (ΔGdiss):
15.54
kcal/mol
Dissociation entropy (TΔSdiss):
13.1
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-184665
Assembly Name:
Purine nucleoside phosphorylase
Multimeric state:
homo dimer
Accessible surface area:
19972.48 Å2
Buried surface area:
5848.69 Å2
Dissociation area:
1,339.1
Å2
Dissociation energy (ΔGdiss):
15.33
kcal/mol
Dissociation entropy (TΔSdiss):
13.09
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-184665
Assembly Name:
Purine nucleoside phosphorylase
Multimeric state:
homo dimer
Accessible surface area:
18152.77 Å2
Buried surface area:
8015.46 Å2
Dissociation area:
2,369.43
Å2
Dissociation energy (ΔGdiss):
4.92
kcal/mol
Dissociation entropy (TΔSdiss):
13.39
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-184665
Assembly Name:
Purine nucleoside phosphorylase
Multimeric state:
homo dimer
Accessible surface area:
18415.69 Å2
Buried surface area:
7011.73 Å2
Dissociation area:
2,281
Å2
Dissociation energy (ΔGdiss):
4.99
kcal/mol
Dissociation entropy (TΔSdiss):
13.36
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-184665
Assembly Name:
Purine nucleoside phosphorylase
Multimeric state:
homo dimer
Accessible surface area:
18173.98 Å2
Buried surface area:
8369.39 Å2
Dissociation area:
2,500.12
Å2
Dissociation energy (ΔGdiss):
-1.53
kcal/mol
Dissociation entropy (TΔSdiss):
13.43
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-184665
Macromolecules
Chains: A, B, C, D, E, F
Length: 276 amino acids
Theoretical weight: 30.5 KDa
Source organism: Plasmodium falciparum 3D7
Expression system: Escherichia coli
UniProt:
Pfam: Phosphorylase superfamily
InterPro:
CATH: Nucleoside phosphorylase domain
SCOP: Purine and uridine phosphorylases
Length: 276 amino acids
Theoretical weight: 30.5 KDa
Source organism: Plasmodium falciparum 3D7
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q8I3X4 (Residues: 2-245; Coverage: 100%)
Q8I3X4 (Residues: 2-245; Coverage: 100%)
Pfam: Phosphorylase superfamily
InterPro:
CATH: Nucleoside phosphorylase domain
SCOP: Purine and uridine phosphorylases