 |
PDBsum entry 4z49
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.1.1.1.100
- 3-oxoacyl-[acyl-carrier-protein] reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (3R)-hydroxyacyl-[ACP] + NADP+ = a 3-oxoacyl-[ACP] + NADPH + H+
|
 |
 |
 |
 |
 |
(3R)-hydroxyacyl-[ACP]
|
+
|
 NADP(+)
NADP(+)
|
=
|
3-oxoacyl-[ACP]
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B:
E.C.1.3.1.39
- enoyl-[acyl-carrier-protein] reductase (Nadph, Re-specific).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2,3-saturated acyl-[ACP] + NADP+ = a (2E)-enoyl-[ACP] + NADPH + H+
|
 |
 |
 |
 |
 |
2,3-saturated acyl-[ACP]
|
+
|
 NADP(+)
NADP(+)
|
=
|
(2E)-enoyl-[ACP]
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B:
E.C.2.3.1.38
- [acyl-carrier-protein] S-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
holo-[ACP] + acetyl-CoA = acetyl-[ACP] + CoA
|
 |
 |
 |
 |
 |
holo-[ACP]
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
acetyl-[ACP]
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains A, B:
E.C.2.3.1.39
- [acyl-carrier-protein] S-malonyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
holo-[ACP] + malonyl-CoA = malonyl-[ACP] + CoA
|
 |
 |
 |
 |
 |
holo-[ACP]
|
+
|
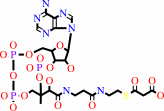 malonyl-CoA
malonyl-CoA
|
=
|
malonyl-[ACP]
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
Chains A, B:
E.C.2.3.1.41
- beta-ketoacyl-[acyl-carrier-protein] synthase I.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a fatty acyl-[ACP] + malonyl-[ACP] + H+ = a 3-oxoacyl-[ACP] + holo- [ACP] + CO2
|
 |
 |
 |
 |
 |
fatty acyl-[ACP]
|
+
|
malonyl-[ACP]
|
+
|
H(+)
|
=
|
3-oxoacyl-[ACP]
|
+
|
holo- [ACP]
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
Chains A, B:
E.C.2.3.1.85
- fatty-acid synthase system.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
acetyl-CoA + n malonyl-CoA + 2n NADPH + 2n H+ = a long-chain fatty acid + (n+1) CoA + n CO2 + 2n NADP+
|
 |
 |
 |
 |
 |
 acetyl-CoA
acetyl-CoA
|
+
|
 n
malonyl-CoA
n
malonyl-CoA
|
+
|
2n NADPH
|
+
|
2n H(+)
|
=
|
 long-chain fatty acid
long-chain fatty acid
|
+
|
(n+1) CoA
|
+
|
 n
CO2
n
CO2
|
+
|
2n NADP(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
Chains A, B:
E.C.3.1.2.14
- oleoyl-[acyl-carrier-protein] hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(9Z)-octadecenoyl-[ACP] + H2O = (9Z)-octadecenoate + holo-[ACP] + H+
|
 |
 |
 |
 |
 |
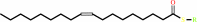 Oleoyl-[acyl-carrier-protein]
Oleoyl-[acyl-carrier-protein]
|
+
|
n
H(2)O
|
=
|
 [acyl-carrier-protein]
[acyl-carrier-protein]
|
+
|
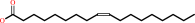 oleate
oleate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 8:
|
 |
Chains A, B:
E.C.4.2.1.59
- 3-hydroxyacyl-[acyl-carrier-protein] dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (3R)-hydroxyacyl-[ACP] = a (2E)-enoyl-[ACP] + H2O
|
 |
 |
 |
 |
 |
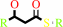 (3R)-3-hydroxyacyl-[acyl-carrier protein]
(3R)-3-hydroxyacyl-[acyl-carrier protein]
|
=
|
n
trans-2-enoyl-[acyl- carrier protein]
|
+
|
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Chem Inf Model
55:1914-1925
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Estimation of Hydrogen-Exchange Protection Factors from MD Simulation Based on Amide Hydrogen Bonding Analysis.
|
|
I.H.Park,
J.D.Venable,
C.Steckler,
S.E.Cellitti,
S.A.Lesley,
G.Spraggon,
A.Brock.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Hydrogen exchange (HX) studies have provided critical insight into our
understanding of protein folding, structure, and dynamics. More recently,
hydrogen exchange mass spectrometry (HX-MS) has become a widely applicable tool
for HX studies. The interpretation of the wealth of data generated by HX-MS
experiments as well as other HX methods would greatly benefit from the
availability of exchange predictions derived from structures or models for
comparison with experiment. Most reported computational HX modeling studies have
employed solvent-accessible-surface-area based metrics in attempts to interpret
HX data on the basis of structures or models. In this study, a computational
HX-MS prediction method based on classification of the amide hydrogen bonding
modes mimicking the local unfolding model is demonstrated. Analysis of the NH
bonding configurations from molecular dynamics (MD) simulation snapshots is used
to determine partitioning over bonded and nonbonded NH states and is directly
mapped into a protection factor (PF) using a logistics growth function.
Predicted PFs are then used for calculating deuteration values of peptides and
compared with experimental data. Hydrogen exchange MS data for fatty acid
synthase thioesterase (FAS-TE) collected for a range of pHs and temperatures was
used for detailed evaluation of the approach. High correlation between
prediction and experiment for observable fragment peptides is observed in the
FAS-TE and additional benchmarking systems that included various apo/holo
proteins for which literature data were available. In addition, it is shown that
HX modeling can improve experimental resolution through decomposition of
in-exchange curves into rate classes, which correlate with prediction from MD.
Successful rate class decompositions provide further evidence that the presented
approach captures the underlying physical processes correctly at the single
residue level. This assessment is further strengthened in a comparison of
residue resolved protection factor predictions for staphylococcal nuclease with
NMR data, which was also used to compare prediction performance with other
algorithms described in the literature. The demonstrated transferable and
scalable MD based HX prediction approach adds significantly to the available
tools for HX-MS data interpretation based on available structures and models.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

