 |
PDBsum entry 1r8c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.72
- Cca tRNA nucleotidyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a tRNA precursor + 2 CTP + ATP = a tRNA with a 3' CCA end + 3 diphosphate
|
 |
 |
 |
 |
 |
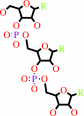 tRNA precursor
tRNA precursor
|
+
|
 2
×
CTP
2
×
CTP
|
+
|
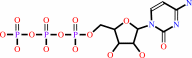
ATP
Bound ligand (Het Group name = )
matches with 93.33% similarity
|
=
|
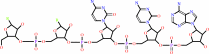 tRNA with a 3' CCA end
tRNA with a 3' CCA end
|
+
|
 3
×
diphosphate
3
×
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Mol Cell
12:1165-1172
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes.
|
|
Y.Xiong,
F.Li,
J.Wang,
A.M.Weiner,
T.A.Steitz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
CCA-adding enzymes catalyze the addition of CCA onto the 3' terminus of immature
tRNAs without using a nucleic acid template and have been divided into two
classes based on their amino acid sequences. We have determined the crystal
structures of a class I CCA-adding enzyme from Archeoglobus fulgidus (AfCCA) and
its complexes with ATP, CTP, or UTP. Although it and the class II bacterial
Bacillus stearothermophilus CCA enzyme (BstCCA) have similar dimensions and
domain architectures (head, neck, body, and tail), only the polymerase domain is
structurally homologous. Moreover, the relative orientation of the head domain
with respect to the body and tail domains, which appear likely to bind tRNA,
differs significantly between the two enzyme classes. Unlike the class II
BstCCA, this enzyme binds nucleotides nonspecifically in the absence of bound
tRNA. The shape and electrostatic charge distribution of the AfCCA enzyme
suggests a model for tRNA binding that accounts for the phosphates that are
protected from chemical modification by tRNA binding to AfCCA. The structures of
the AfCCA enzyme and the eukaryotic poly(A) polymerase are very similar,
implying a close evolutionary relationship between them.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. A Comparison of the Structures of the Polymerase
Domains of AfCCA, BstCCA, and DNA pol βThe polymerase domain is
shown in magenta and the neck/fingers in green. The incoming
nucleotide is yellow and the metal ions are brown. The three
catalytic carboxylates are shown. In (B), the last magenta helix
(cylinder) to the right belongs to pol β fingers domain. For
comparison with the AfCCA head it is colored the same as the
palm domain. In (C), the region colored in red represents the
additional part of BstCCA not seen in the AfCCA head and Pol β
domains.
|
 |
Figure 5.
Figure 5. A Comparison of the Different Orientations that
the tRNA Acceptor Stem and TΨC Stem-Loop Have with Respect to
the Head Domains in the Two Classes of CCA-Adding Enzyme(A)
AfCCA. (B) BstCCA. The head domains of the two enzymes are
represented in magenta ribbons and are in the same orientation.
The enzymes are shown with surface representation and the
acceptor stem and TψC stem-loop of the tRNA in yellow coil.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Mol Cell
(2003,
12,
1165-1172)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.Pan,
Y.Xiong,
and
T.A.Steitz
(2010).
How the CCA-adding enzyme selects adenine over cytosine at position 76 of tRNA.
|
| |
Science,
330,
937-940.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.U.Heinemann,
D.Söll,
and
L.Randau
(2010).
Transfer RNA processing in archaea: unusual pathways and enzymes.
|
| |
FEBS Lett,
584,
303-309.
|
 |
|
|
|
|
 |
I.Y.Morozov,
M.G.Jones,
A.A.Razak,
D.J.Rigden,
and
M.X.Caddick
(2010).
CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans.
|
| |
Mol Cell Biol,
30,
460-469.
|
 |
|
|
|
|
 |
S.J.Hyde,
B.E.Eckenroth,
B.A.Smith,
W.A.Eberley,
N.H.Heintz,
J.E.Jackman,
and
S.Doublié
(2010).
tRNA(His) guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases.
|
| |
Proc Natl Acad Sci U S A,
107,
20305-20310.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.M.Hou
(2010).
CCA addition to tRNA: implications for tRNA quality control.
|
| |
IUBMB Life,
62,
251-260.
|
 |
|
|
|
|
 |
S.Kim,
C.Liu,
K.Halkidis,
H.B.Gamper,
and
Y.M.Hou
(2009).
Distinct kinetic determinants for the stepwise CCA addition to tRNA.
|
| |
RNA,
15,
1827-1836.
|
 |
|
|
|
|
 |
Y.Toh,
D.Takeshita,
T.Numata,
S.Fukai,
O.Nureki,
and
K.Tomita
(2009).
Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme.
|
| |
EMBO J,
28,
3353-3365.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Martin,
S.Doublié,
and
W.Keller
(2008).
Determinants of substrate specificity in RNA-dependent nucleotidyl transferases.
|
| |
Biochim Biophys Acta,
1779,
206-216.
|
 |
|
|
|
|
 |
J.J.Ellis,
and
S.Jones
(2008).
Evaluating conformational changes in protein structures binding RNA.
|
| |
Proteins,
70,
1518-1526.
|
 |
|
|
|
|
 |
M.Dupasquier,
S.Kim,
K.Halkidis,
H.Gamper,
and
Y.M.Hou
(2008).
tRNA integrity is a prerequisite for rapid CCA addition: implication for quality control.
|
| |
J Mol Biol,
379,
579-588.
|
 |
|
|
|
|
 |
X.Shan,
T.A.Russell,
S.M.Paul,
D.B.Kushner,
and
P.B.Joyce
(2008).
Characterization of a temperature-sensitive mutation that impairs the function of yeast tRNA nucleotidyltransferase.
|
| |
Yeast,
25,
219-233.
|
 |
|
|
|
|
 |
Y.Toh,
T.Numata,
K.Watanabe,
D.Takeshita,
O.Nureki,
and
K.Tomita
(2008).
Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme.
|
| |
EMBO J,
27,
1944-1952.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Gao,
and
R.S.Gupta
(2007).
Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis.
|
| |
BMC Genomics,
8,
86.
|
 |
|
|
|
|
 |
G.Martin,
and
W.Keller
(2007).
RNA-specific ribonucleotidyl transferases.
|
| |
RNA,
13,
1834-1849.
|
 |
|
|
|
|
 |
H.D.Cho,
Y.Chen,
G.Varani,
and
A.M.Weiner
(2006).
A model for C74 addition by CCA-adding enzymes: C74 addition, like C75 and A76 addition, does not involve tRNA translocation.
|
| |
J Biol Chem,
281,
9801-9811.
|
 |
|
|
|
|
 |
J.E.Jackman,
and
E.M.Phizicky
(2006).
tRNAHis guanylyltransferase catalyzes a 3'-5' polymerization reaction that is distinct from G-1 addition.
|
| |
Proc Natl Acad Sci U S A,
103,
8640-8645.
|
 |
|
|
|
|
 |
K.Tomita,
R.Ishitani,
S.Fukai,
and
O.Nureki
(2006).
Complete crystallographic analysis of the dynamics of CCA sequence addition.
|
| |
Nature,
443,
956-960.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.D.Cho,
C.L.Verlinde,
and
A.M.Weiner
(2005).
Archaeal CCA-adding enzymes: central role of a highly conserved beta-turn motif in RNA polymerization without translocation.
|
| |
J Biol Chem,
280,
9555-9566.
|
 |
|
|
|
|
 |
J.Deng,
N.L.Ernst,
S.Turley,
K.D.Stuart,
and
W.G.Hol
(2005).
Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
|
| |
EMBO J,
24,
4007-4017.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Randau,
R.Münch,
M.J.Hohn,
D.Jahn,
and
D.Söll
(2005).
Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5'- and 3'-halves.
|
| |
Nature,
433,
537-541.
|
 |
|
|
|
|
 |
A.M.Weiner
(2004).
tRNA maturation: RNA polymerization without a nucleic acid template.
|
| |
Curr Biol,
14,
R883-R885.
|
 |
|
|
|
|
 |
G.Martin,
and
W.Keller
(2004).
Sequence motifs that distinguish ATP(CTP):tRNA nucleotidyl transferases from eubacterial poly(A) polymerases.
|
| |
RNA,
10,
899-906.
|
 |
|
|
|
|
 |
H.D.Cho,
and
A.M.Weiner
(2004).
A single catalytically active subunit in the multimeric Sulfolobus shibatae CCA-adding enzyme can carry out all three steps of CCA addition.
|
| |
J Biol Chem,
279,
40130-40136.
|
 |
|
|
|
|
 |
K.Tomita,
S.Fukai,
R.Ishitani,
T.Ueda,
N.Takeuchi,
D.G.Vassylyev,
and
O.Nureki
(2004).
Structural basis for template-independent RNA polymerization.
|
| |
Nature,
430,
700-704.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Schimmel,
and
X.L.Yang
(2004).
Two classes give lessons about CCA.
|
| |
Nat Struct Mol Biol,
11,
807-808.
|
 |
|
|
|
|
 |
Y.Xiong,
and
T.A.Steitz
(2004).
Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template.
|
| |
Nature,
430,
640-645.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

