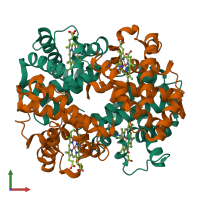
Assemblies
Assembly Name:
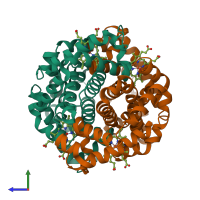
Hemoglobin HbA complex
Multimeric state:
hetero tetramer
Accessible surface area:
23501.68 Å2
Buried surface area:
11036.78 Å2
Dissociation area:
1,395.5
Å2
Dissociation energy (ΔGdiss):
5.46
kcal/mol
Dissociation entropy (TΔSdiss):
13.65
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-159519
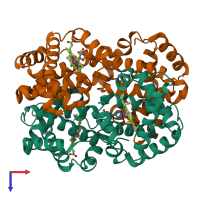
Assembly Name:
Hemoglobin HbA complex
Multimeric state:
hetero tetramer
Accessible surface area:
24087.76 Å2
Buried surface area:
10774.44 Å2
Dissociation area:
3,065.32
Å2
Dissociation energy (ΔGdiss):
5.66
kcal/mol
Dissociation entropy (TΔSdiss):
36.68
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-159519
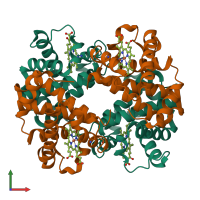
Assembly Name:
Hemoglobin HbA complex
Multimeric state:
hetero tetramer
Accessible surface area:
23942.74 Å2
Buried surface area:
11011.89 Å2
Dissociation area:
3,187.7
Å2
Dissociation energy (ΔGdiss):
4.81
kcal/mol
Dissociation entropy (TΔSdiss):
36.71
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-159519
Macromolecules
Chains: B, D, F, H, J, L
Length: 146 amino acids
Theoretical weight: 15.89 KDa
Source organism: Homo sapiens
UniProt:
Pfam: Globin
InterPro:
CATH: Globins
Length: 146 amino acids
Theoretical weight: 15.89 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P68871 (Residues: 2-147; Coverage: 99%)
P68871 (Residues: 2-147; Coverage: 99%)
Pfam: Globin
InterPro:
CATH: Globins