Assemblies
Assembly Name:
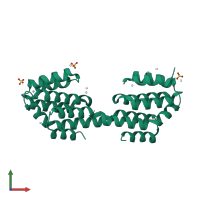
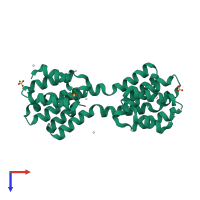
Regulation of nuclear pre-mRNA domain-containing protein 1B
Multimeric state:
homo dimer
Accessible surface area:
13457.33 Å2
Buried surface area:
3621.8 Å2
Dissociation area:
1,586.97
Å2
Dissociation energy (ΔGdiss):
23.84
kcal/mol
Dissociation entropy (TΔSdiss):
12.35
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-192503
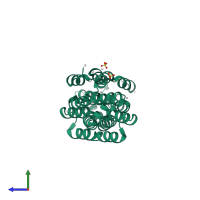
Assembly Name:
Regulation of nuclear pre-mRNA domain-containing protein 1B
Multimeric state:
homo dimer
Accessible surface area:
13601.86 Å2
Buried surface area:
3432.1 Å2
Dissociation area:
1,645.26
Å2
Dissociation energy (ΔGdiss):
24.1
kcal/mol
Dissociation entropy (TΔSdiss):
12.38
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-192503
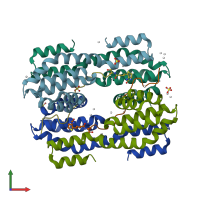
Assembly Name:
Regulation of nuclear pre-mRNA domain-containing protein 1B
Multimeric state:
hetero hexamer
Accessible surface area:
24953.93 Å2
Buried surface area:
14808.86 Å2
Dissociation area:
3,460.9
Å2
Dissociation energy (ΔGdiss):
28.43
kcal/mol
Dissociation entropy (TΔSdiss):
19.44
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-165083
Macromolecules
Chains: A, B, D, E
Length: 135 amino acids
Theoretical weight: 15.56 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: CID domain
InterPro:
CATH: Serine Threonine Protein Phosphatase 5, Tetratricopeptide repeat
Length: 135 amino acids
Theoretical weight: 15.56 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q9NQG5 (Residues: 2-135; Coverage: 41%)
Q9NQG5 (Residues: 2-135; Coverage: 41%)
Pfam: CID domain
InterPro:
CATH: Serine Threonine Protein Phosphatase 5, Tetratricopeptide repeat
Chains: C, F
Length: 21 amino acids
Theoretical weight: 2.2 KDa
Source organism: Homo sapiens
Expression system: Not provided
Length: 21 amino acids
Theoretical weight: 2.2 KDa
Source organism: Homo sapiens
Expression system: Not provided