Assemblies
Multimeric state:
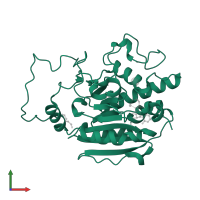
monomeric
Accessible surface area:
13899.39 Å2
Buried surface area:
1472.18 Å2
Dissociation area:
131.65
Å2
Dissociation energy (ΔGdiss):
-3.87
kcal/mol
Dissociation entropy (TΔSdiss):
1.28
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-147725
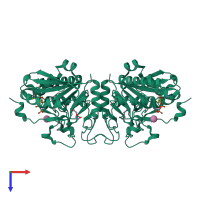
Multimeric state:
homo dimer
Accessible surface area:
22686.81 Å2
Buried surface area:
8053.82 Å2
Dissociation area:
2,553.04
Å2
Dissociation energy (ΔGdiss):
23.03
kcal/mol
Dissociation entropy (TΔSdiss):
13.88
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-147726
Macromolecules
Chain: A
Length: 293 amino acids
Theoretical weight: 34.19 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21
UniProt:
Pfam: Glycosyltransferase family 6
InterPro:
CATH: Spore Coat Polysaccharide Biosynthesis Protein SpsA; Chain A
Length: 293 amino acids
Theoretical weight: 34.19 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21
UniProt:
- Canonical:
 P16442 (Residues: 64-354; Coverage: 82%)
P16442 (Residues: 64-354; Coverage: 82%)
Pfam: Glycosyltransferase family 6
InterPro:
CATH: Spore Coat Polysaccharide Biosynthesis Protein SpsA; Chain A