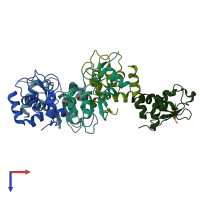
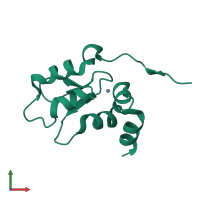
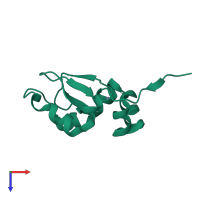
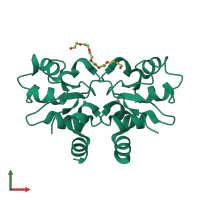
Assemblies
Assembly Name:
Baculoviral IAP repeat-containing protein 7 30kDa subunit
Multimeric state:
monomeric
Accessible surface area:
6271.59 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-188400
Assembly Name:
Baculoviral IAP repeat-containing protein 7 30kDa subunit
Multimeric state:
homo dimer
Accessible surface area:
10397.95 Å2
Buried surface area:
2672.5 Å2
Dissociation area:
1,181.83
Å2
Dissociation energy (ΔGdiss):
2.82
kcal/mol
Dissociation entropy (TΔSdiss):
11.3
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-188402
Assembly Name:
Baculoviral IAP repeat-containing protein 7 30kDa subunit
Multimeric state:
monomeric
Accessible surface area:
5833.35 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-188400
Assembly Name:
Short peptide
Multimeric state:
monomeric
Accessible surface area:
620.27 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-219427
Assembly Name:
Baculoviral IAP repeat-containing protein 7 30kDa subunit
Multimeric state:
monomeric
Accessible surface area:
6426.06 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-188400
Assembly Name:
Baculoviral IAP repeat-containing protein 7 30kDa subunit
Multimeric state:
monomeric
Accessible surface area:
6469.83 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-188400
Assembly Name:
Baculoviral IAP repeat-containing protein 7 30kDa subunit
Multimeric state:
monomeric
Accessible surface area:
6587.12 Å2
Buried surface area:
309.56 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-188400
Multimeric state:
hetero dimer
Accessible surface area:
5718.0 Å2
Buried surface area:
637.81 Å2
Dissociation area:
318.9
Å2
Dissociation energy (ΔGdiss):
0.94
kcal/mol
Dissociation entropy (TΔSdiss):
4.6
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-162367
Multimeric state:
hetero hexamer
Accessible surface area:
23766.07 Å2
Buried surface area:
8364.39 Å2
Dissociation area:
830.37
Å2
Dissociation energy (ΔGdiss):
-1.34
kcal/mol
Dissociation entropy (TΔSdiss):
15.75
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-162370
Multimeric state:
hetero tetramer
Accessible surface area:
15096.97 Å2
Buried surface area:
4333.24 Å2
Dissociation area:
830.37
Å2
Dissociation energy (ΔGdiss):
-0.89
kcal/mol
Dissociation entropy (TΔSdiss):
15.29
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-162369
Assembly Name:
Baculoviral IAP repeat-containing protein 7 30kDa subunit
Multimeric state:
homo dimer
Accessible surface area:
10659.65 Å2
Buried surface area:
2040.6 Å2
Dissociation area:
1,020.3
Å2
Dissociation energy (ΔGdiss):
-0.05
kcal/mol
Dissociation entropy (TΔSdiss):
11.24
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-188402
Multimeric state:
hetero trimer
Accessible surface area:
11286.14 Å2
Buried surface area:
1970.3 Å2
Dissociation area:
830.37
Å2
Dissociation energy (ΔGdiss):
-0.74
kcal/mol
Dissociation entropy (TΔSdiss):
15.14
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-162368
Macromolecules
Chains: A, B, C, D, E
Length: 140 amino acids
Theoretical weight: 15.75 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Inhibitor of Apoptosis domain
InterPro: BIR repeat
CATH: Inhibitor Of Apoptosis Protein (2mihbC-IAP-1); Chain A
SCOP: Inhibitor of apoptosis (IAP) repeat
Length: 140 amino acids
Theoretical weight: 15.75 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q96CA5 (Residues: 63-179; Coverage: 39%)
Q96CA5 (Residues: 63-179; Coverage: 39%)
Pfam: Inhibitor of Apoptosis domain
InterPro: BIR repeat
CATH: Inhibitor Of Apoptosis Protein (2mihbC-IAP-1); Chain A
SCOP: Inhibitor of apoptosis (IAP) repeat
Chain: F
Length: 9 amino acids
Theoretical weight: 1.02 KDa
Source organism: Homo sapiens
Expression system: Not provided
Length: 9 amino acids
Theoretical weight: 1.02 KDa
Source organism: Homo sapiens
Expression system: Not provided