Assemblies
Multimeric state:
hetero dimer
Accessible surface area:
15188.35 Å2
Buried surface area:
1392.4 Å2
Dissociation area:
696.2
Å2
Dissociation energy (ΔGdiss):
2.29
kcal/mol
Dissociation entropy (TΔSdiss):
7.62
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-152088
Multimeric state:
hetero dimer
Accessible surface area:
15487.46 Å2
Buried surface area:
1405.77 Å2
Dissociation area:
702.89
Å2
Dissociation energy (ΔGdiss):
1.05
kcal/mol
Dissociation entropy (TΔSdiss):
7.62
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-152088
Macromolecules
Chains: A, B
Length: 335 amino acids
Theoretical weight: 39.07 KDa
Source organism: Homo sapiens
Expression system: Spodoptera frugiperda
UniProt:
Pfam:
InterPro:
SCOP: Protein kinases, catalytic subunit
Length: 335 amino acids
Theoretical weight: 39.07 KDa
Source organism: Homo sapiens
Expression system: Spodoptera frugiperda
UniProt:
- Canonical:
 P31751 (Residues: 146-480; Coverage: 70%)
P31751 (Residues: 146-480; Coverage: 70%)
Pfam:
InterPro:
- Protein kinase-like domain superfamily
- Protein kinase domain
- Protein Kinase B beta, catalytic domain
- Protein kinase, ATP binding site
- Serine/threonine-protein kinase, active site
- AGC-kinase, C-terminal
- Protein kinase, C-terminal
SCOP: Protein kinases, catalytic subunit
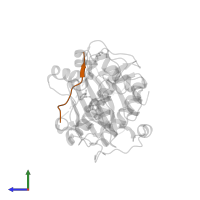
Chains: C, D
Length: 10 amino acids
Theoretical weight: 1.12 KDa
Source organism: Homo sapiens
Expression system: Not provided
UniProt:
Length: 10 amino acids
Theoretical weight: 1.12 KDa
Source organism: Homo sapiens
Expression system: Not provided
UniProt:
- Canonical:
 P49841 (Residues: 3-12; Coverage: 2%)
P49841 (Residues: 3-12; Coverage: 2%)



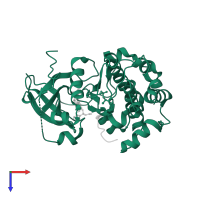
![The deposited structure of PDB entry <span class='highlight'>3e8d</span> coloured by chain, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 3e8d coloured by chain, front view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_deposited_chain_front_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>3e8d</span> coloured by chain, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 3e8d coloured by chain, side view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_deposited_chain_side_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>3e8d</span> coloured by chain, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 3e8d coloured by chain, top view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_deposited_chain_top_image-200x200.png)
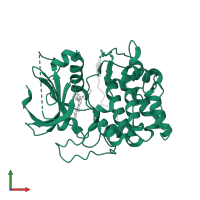
![Hetero dimeric assembly 1 of PDB entry <span class='highlight'>3e8d</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero dimeric assembly 1 of PDB entry 3e8d coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_assembly_1_chemically_distinct_molecules_front_image-200x200.png)
![Hetero dimeric assembly 1 of PDB entry <span class='highlight'>3e8d</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero dimeric assembly 1 of PDB entry 3e8d coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_assembly_1_chemically_distinct_molecules_side_image-200x200.png)
![Hetero dimeric assembly 1 of PDB entry <span class='highlight'>3e8d</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero dimeric assembly 1 of PDB entry 3e8d coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_assembly_1_chemically_distinct_molecules_top_image-200x200.png)
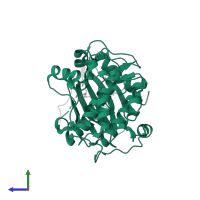
![Hetero dimeric assembly 2 of PDB entry <span class='highlight'>3e8d</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero dimeric assembly 2 of PDB entry 3e8d coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_assembly_2_chemically_distinct_molecules_front_image-200x200.png)
![Hetero dimeric assembly 2 of PDB entry <span class='highlight'>3e8d</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero dimeric assembly 2 of PDB entry 3e8d coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_assembly_2_chemically_distinct_molecules_side_image-200x200.png)
![Hetero dimeric assembly 2 of PDB entry <span class='highlight'>3e8d</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>RAC-beta serine/threonine-protein kinase</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Glycogen synthase kinase-3 beta</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero dimeric assembly 2 of PDB entry 3e8d coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/3e8d_assembly_2_chemically_distinct_molecules_top_image-200x200.png)