Epoxide hydrolase
Epoxide hydrolases convert epoxides into more soluble and less toxic products, an essential process within cells. As a result, these enzymes have been found in a wide variety of organisms. Their ability to react enantioselectively with industrially important epoxides makes epoxide hydrolases promising bio-catalysts for the preparation of enantiomerically pure epoxides, and/or their corresponding vicinal diols.
Reference Protein and Structure
- Sequence
-
O31243
 (3.3.2.3)
(3.3.2.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Agrobacterium tumefaciens (Bacteria)

- PDB
-
1ehy
- X-ray structure of the epoxide hydrolase from agrobacterium radiobacter ad1
(2.1 Å)



- Catalytic CATH Domains
-
3.40.50.1820
 (see all for 1ehy)
(see all for 1ehy)
Enzyme Reaction (EC:3.3.2.10)
Enzyme Mechanism
Introduction
The mechanism involves two steps. In the first reaction an ester bond is formed between the enzyme and substrate by attack of the nucleophilic Asp 107 on the primary carbon of the substrate; in the second step this ester bond is hydrolysed by a water molecule, activated by the His275/Asp246 pair. The anionic tetrahedral intermediate is stabilised by hydrogen bond interactions with the backbone amide nitrogens of Phe 108 and Trp 38.
Catalytic Residues Roles
| UniProt | PDB* (1ehy) | ||
| Tyr152 | Tyr152A | This residue is less well conserved that Tyr215, but also forms a hydrogen bond with the epoxide oxygen so could also act as the general acid/base. However, it is more likely to help stabilise the negatively charged Tyr215 that results from the removal of its proton. | increase basicity, electrostatic stabiliser |
| Asp107 | Asp107A | Acts as a nucleophile, forming a covalent bond with the epoxide substrate. | covalently attached, nucleophile, increase acidity, electrofuge, electrophile |
| His275 | His275A | Acts as a general acid/base. Part of a catalytic dyad with Asp246. | proton acceptor, proton donor |
| Phe108 (main-N), Trp38 (main-N) | Phe108A (main-N), Trp38A (main-N) | Helps activate the nucleophilic Asp side chain. Helps stabilise the reactive intermediates and transition states. | electrostatic stabiliser |
| Asp246 | Asp246A | Activates the general acid/base His275. | increase basicity, electrostatic stabiliser |
| Tyr215 | Tyr215A | Possible general acid/base that protonates the epoxide oxygen during the step in which the nucleophilic aspartate forms a covalent bond with the substrate. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, enzyme-substrate complex cleavage, overall product formed, unimolecular elimination by the conjugate base, inferred reaction step, native state of enzyme regeneratedReferences
- Nardini M et al. (1999), J Biol Chem, 274, 14579-14586. The X-ray Structure of Epoxide Hydrolase from Agrobacterium radiobacter AD1: AN ENZYME TO DETOXIFY HARMFUL EPOXIDES. DOI:10.1074/jbc.274.21.14579. PMID:10329649.
- Lonsdale R et al. (2012), Biochemistry, 51, 1774-1786. Determinants of reactivity and selectivity in soluble epoxide hydrolase from quantum mechanics/molecular mechanics modeling. DOI:10.1021/bi201722j. PMID:22280021.
- Blée E et al. (2005), J Biol Chem, 280, 6479-6487. Soybean epoxide hydrolase: identification of the catalytic residues and probing of the reaction mechanism with secondary kinetic isotope effects. DOI:10.1074/jbc.M411366200. PMID:15596432.

Step 1. Asp107 initiates a nucleophilic attack on the epoxide, forming an covalently bound intermediate with concommittant abstraction of a proton from Tyr215.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Trp38A (main-N) | electrostatic stabiliser |
| Phe108A (main-N) | electrostatic stabiliser |
| Tyr152A | electrostatic stabiliser |
| Asp107A | nucleophile |
| Tyr215A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation
Step 2. His275 abstracts a proton from the substrate water, which attacks the bound aspartate residue.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Trp38A (main-N) | electrostatic stabiliser |
| Phe108A (main-N) | electrostatic stabiliser |
| Tyr152A | electrostatic stabiliser |
| Asp246A | increase basicity |
| Asp107A | covalently attached |
| His275A | proton acceptor |
| Asp107A | electrophile |
Chemical Components
proton transfer, overall reactant used, ingold: bimolecular nucleophilic addition
Step 3. The oxyanion collapses, eliminating the product and re-forming the asparate residue.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Trp38A (main-N) | electrostatic stabiliser |
| Phe108A (main-N) | electrostatic stabiliser |
| Tyr152A | electrostatic stabiliser |
| Asp246A | electrostatic stabiliser |
| Asp107A | electrofuge |



 Download:
Download: 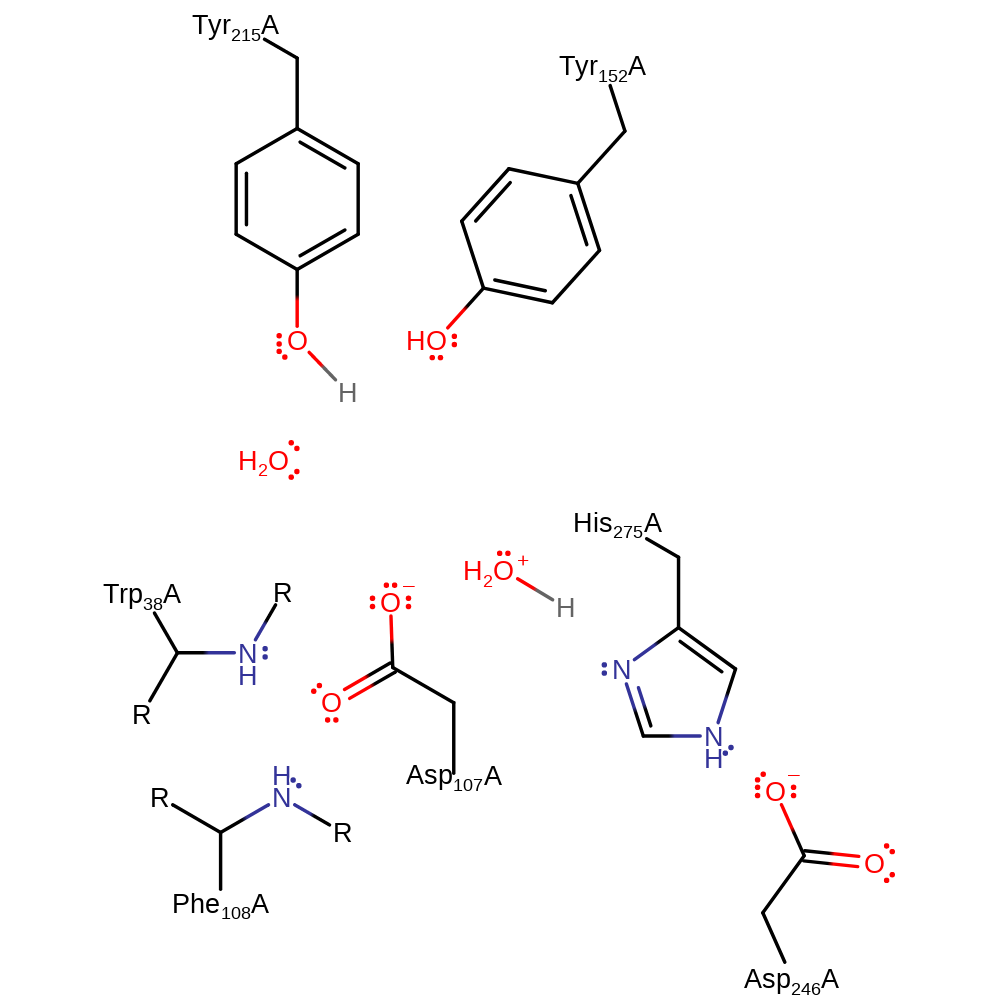 Download:
Download: