DUTP diphosphatase
dUTP diphosphatase(dUTPase) hydrolyses dUTP to dUMP and pyrophosphate, simultaneously reducing dUTP levels and providing the dUMP for dTTP biosynthesis. This reaction is critical for the fidelity of DNA replication and repair as dUTPase decreases the intracellular concentration of dUPT so that uracil cannot be incorporated into DNA.
Reference Protein and Structure
- Sequence
-
P06968
 (3.6.1.23)
(3.6.1.23)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1dup
- DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE (D-UTPASE)
(1.9 Å)



- Catalytic CATH Domains
-
2.70.40.10
 (see all for 1dup)
(see all for 1dup)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:3.6.1.23)
+
→
+
+
Alternative enzyme names: dUTP pyrophosphatase, dUTPase, Deoxyuridine-triphosphatase, Desoxyuridine 5'-triphosphatase, Desoxyuridine 5'-triphosphate nucleotidohydrolase,
Enzyme Mechanism
Introduction
Asp90 activates the catalytic water, which attacks the gamma phosphate, elimintating the diphoshate and forming dUMP.
Catalytic Residues Roles
| UniProt | PDB* (1dup) | ||
| Ala28 (main-N) | Ala29A (main-N) | Hydrogen bonds to Asp90 side chain, helping to hold it in the correct orientation and activating it to act as a general acid/base. | activator |
| Gly72 (main-N), Arg70 | Gly73A (main-N), Arg71A | Along with Arg141 (in the disorderd C-terminal Motif V), these residues participate in pyrophosphate escape via charge stabilisation. | electrostatic stabiliser |
| Ile79 (main-C) | Ile80A (main-C) | Hydrogen bonds to the catalytic water, activating it for nucleophilic attack. | modifies pKa |
| Asp89 | Asp90A(AA) | Acts as a general acid/base, activating the catalytic water. | proton acceptor, proton donor |
*PDB label guide - RESx(y)B(C) - RES: Residue Name; x: Residue ID in PDB file;
y: Residue ID in PDB sequence if different from PDB file; B: PDB Chain;
C: Biological Assembly Chain if different from PDB. If label is "Not Found" it means this residue is not found in the reference PDB.
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, overall product formed, dephosphorylation, hydrolysis, inferred reaction step, native state of enzyme regeneratedReferences
- Barabás O et al. (2004), J Biol Chem, 279, 42907-42915. Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase. DOI:10.1074/jbc.M406135200. PMID:15208312.
- Barabás O et al. (2013), Nucleic Acids Res, 41, 10542-10555. Catalytic mechanism of α-phosphate attack in dUTPase is revealed by X-ray crystallographic snapshots of distinct intermediates, 31P-NMR spectroscopy and reaction path modelling. DOI:10.1093/nar/gkt756. PMID:23982515.
- Cedergren-Zeppezauer ES et al. (1992), Nature, 355, 740-743. Crystal structure of a dUTPase. DOI:10.1038/355740a0. PMID:1311056.
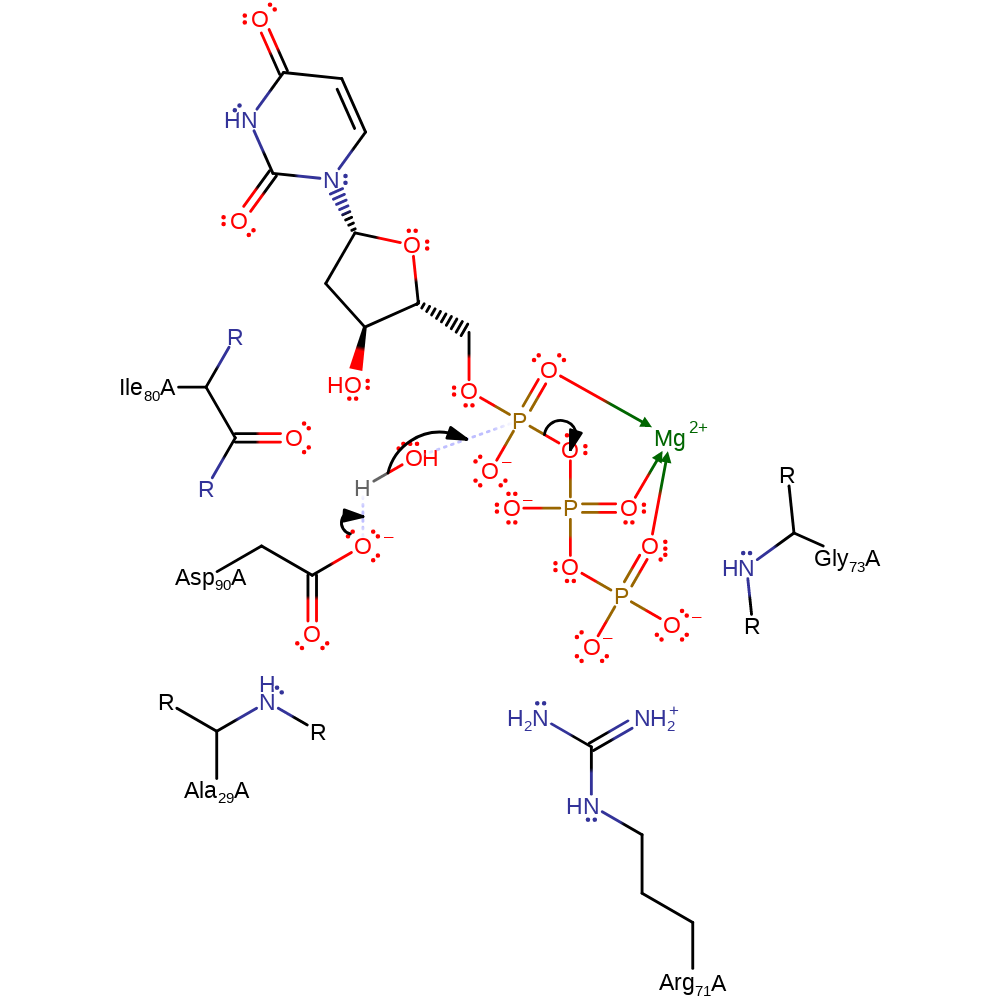
Step 1. Asp90 abstracts a proton from water, which initiates a nucleophilic attack on the gamma phosphate, eliminating pyrophosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg71A | electrostatic stabiliser |
| Gly73A (main-N) | electrostatic stabiliser |
| Ile80A (main-C) | modifies pKa |
| Ala29A (main-N) | activator |
| Asp90A(AA) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, dephosphorylation, hydrolysis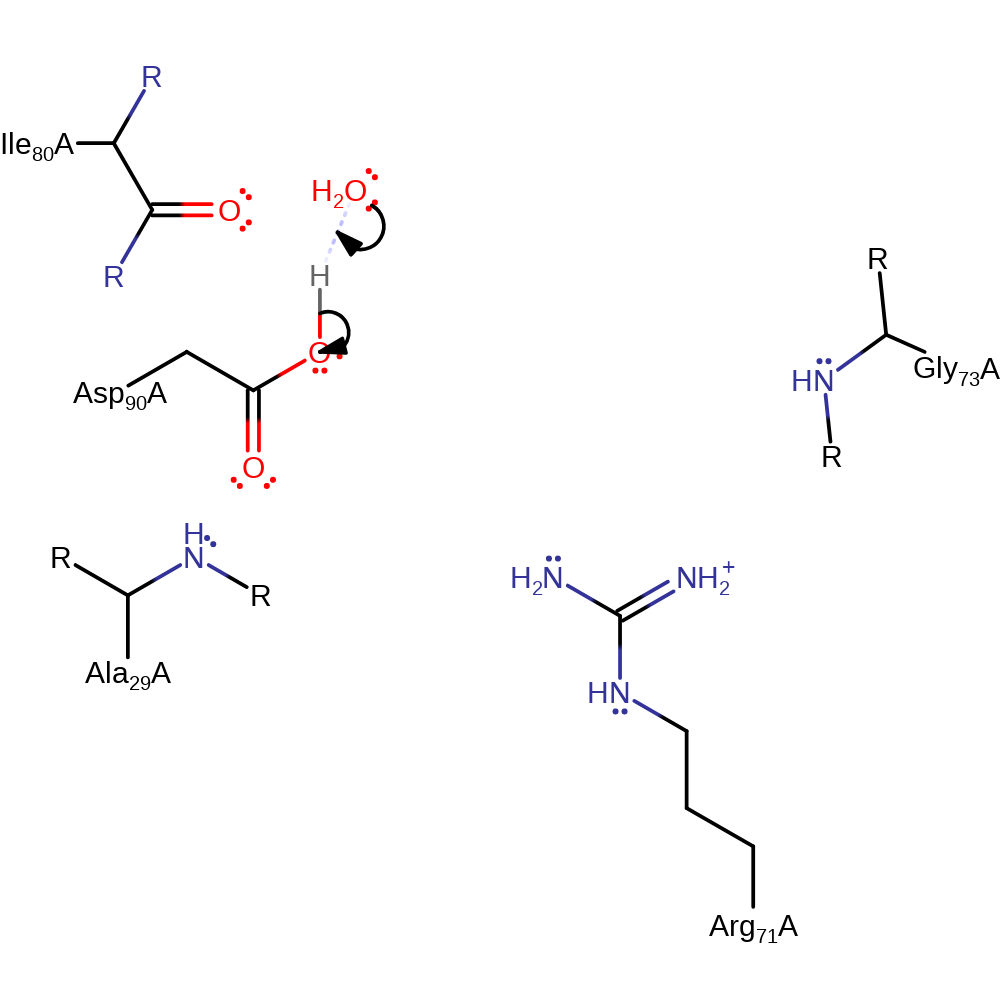
Step 2. In an inferred return step, a water molecule abstracts a proton from Asp90, regenerating the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ala29A (main-N) | activator |
| Ile80A (main-C) | modifies pKa |
| Asp90A(AA) | proton donor |





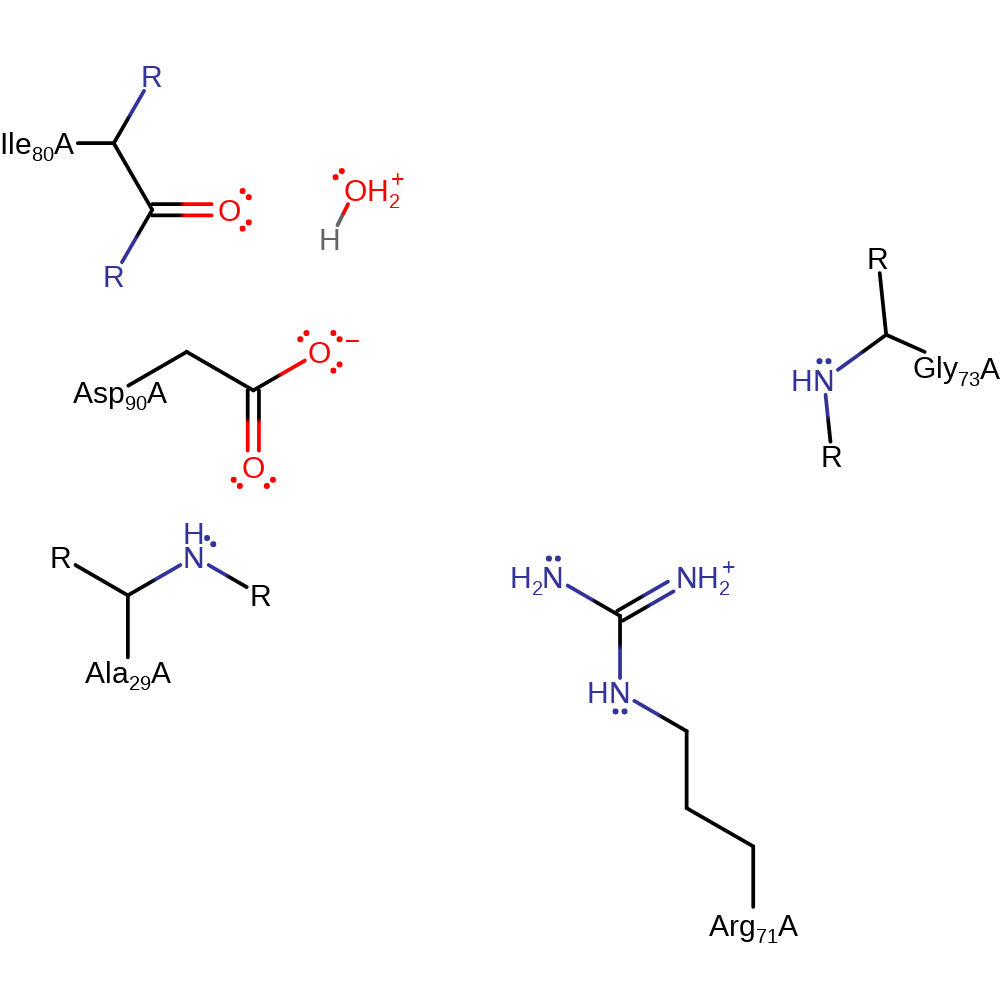 Download:
Download: