 |
PDBsum entry 1dup

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.6.1.23
- dUTP diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dUTP + H2O = dUMP + diphosphate + H+
|
 |
 |
 |
 |
 |
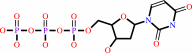 dUTP
dUTP
|
+
|
H2O
|
=
|
 dUMP
dUMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nature
355:740-743
(1992)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a dUTPase.
|
|
E.S.Cedergren-Zeppezauer,
G.Larsson,
P.O.Nyman,
Z.Dauter,
K.S.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The enzyme dUTPase catalyses the hydrolysis of dUTP and maintains a low
intracellular concentration of dUTP so that uracil cannot be incorporated into
DNA. dUTPase from Escherichia coli is strictly specific for its dUTP substrate,
the active site discriminating between nucleotides with respect to the sugar
moiety as well as the pyrimidine base. Here we report the three-dimensional
structure of E. coli dUTPase determined by X-ray crystallography at a resolution
of 1.9 A. The enzyme is a symmetrical trimer, and of the 152 amino acid residues
in the subunit, the first 136 are visible in the crystal structure. The tertiary
structure resembles a jelly-roll fold and does not show the 'classical'
nucleotide-binding domain. In the quaternary structure there is a complex
interaction between the subunits that may be important in catalysis. This
possibility is supported by the location of conserved elements in the sequence.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.W.Han,
M.A.Elsliger,
T.O.Yeates,
Q.Xu,
A.G.Murzin,
S.S.Krishna,
L.Jaroszewski,
P.Abdubek,
T.Astakhova,
H.L.Axelrod,
D.Carlton,
C.Chen,
H.J.Chiu,
T.Clayton,
D.Das,
M.C.Deller,
L.Duan,
D.Ernst,
J.Feuerhelm,
J.C.Grant,
A.Grzechnik,
K.K.Jin,
H.A.Johnson,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
A.Kumar,
W.W.Lam,
D.Marciano,
D.McMullan,
M.D.Miller,
A.T.Morse,
E.Nigoghossian,
L.Okach,
R.Reyes,
C.L.Rife,
N.Sefcovic,
H.J.Tien,
C.B.Trame,
H.van den Bedem,
D.Weekes,
K.O.Hodgson,
J.Wooley,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
and
I.A.Wilson
(2010).
Structure of a putative NTP pyrophosphohydrolase: YP_001813558.1 from Exiguobacterium sibiricum 255-15.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1237-1244.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.García-Nafría,
L.Burchell,
M.Takezawa,
N.J.Rzechorzek,
M.J.Fogg,
and
K.S.Wilson
(2010).
The structure of the genomic Bacillus subtilis dUTPase: novel features in the Phe-lid.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
953-961.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Homma,
and
H.Moriyama
(2009).
Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
1030-1034.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Freeman,
M.Buisson,
N.Tarbouriech,
A.Van der Heyden,
P.Labbé,
and
W.P.Burmeister
(2009).
The flexible motif V of Epstein-Barr virus deoxyuridine 5'-triphosphate pyrophosphatase is essential for catalysis.
|
| |
J Biol Chem,
284,
25280-25289.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Johansson,
M.Thymark,
J.H.Bynck,
M.Fanø,
S.Larsen,
and
M.Willemoës
(2007).
Regulation of dCTP deaminase from Escherichia coli by nonallosteric dTTP binding to an inactive form of the enzyme.
|
| |
FEBS J,
274,
4188-4198.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Bajaj,
and
H.Moriyama
(2007).
Purification, crystallization and preliminary crystallographic analysis of deoxyuridine triphosphate nucleotidohydrolase from Arabidopsis thaliana.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
409-411.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Cho,
H.S.Lee,
Y.J.Kim,
S.G.Kang,
S.J.Kim,
and
J.H.Lee
(2007).
Characterization of a dUTPase from the hyperthermophilic archaeon Thermococcus onnurineus NA1 and its application in polymerase chain reaction amplification.
|
| |
Mar Biotechnol (NY),
9,
450-458.
|
 |
|
|
|
|
 |
A.J.Davison,
and
N.D.Stow
(2005).
New genes from old: redeployment of dUTPase by herpesviruses.
|
| |
J Virol,
79,
12880-12892.
|
 |
|
|
|
|
 |
J.L.Whittingham,
I.Leal,
C.Nguyen,
G.Kasinathan,
E.Bell,
A.F.Jones,
C.Berry,
A.Benito,
J.P.Turkenburg,
E.J.Dodson,
L.M.Ruiz Perez,
A.J.Wilkinson,
N.G.Johansson,
R.Brun,
I.H.Gilbert,
D.Gonzalez Pacanowska,
and
K.S.Wilson
(2005).
dUTPase as a platform for antimalarial drug design: structural basis for the selectivity of a class of nucleoside inhibitors.
|
| |
Structure,
13,
329-338.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Tarbouriech,
M.Buisson,
J.M.Seigneurin,
S.Cusack,
and
W.P.Burmeister
(2005).
The monomeric dUTPase from Epstein-Barr virus mimics trimeric dUTPases.
|
| |
Structure,
13,
1299-1310.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Zhang,
H.Moriyama,
K.Homma,
and
J.L.Van Etten
(2005).
Chlorella virus-encoded deoxyuridine triphosphatases exhibit different temperature optima.
|
| |
J Virol,
79,
9945-9953.
|
 |
|
|
|
|
 |
E.A.Kouzminova,
and
A.Kuzminov
(2004).
Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair.
|
| |
Mol Microbiol,
51,
1279-1295.
|
 |
|
|
|
|
 |
J.Kovári,
O.Barabás,
E.Takács,
A.Békési,
Z.Dubrovay,
V.Pongrácz,
I.Zagyva,
T.Imre,
P.Szabó,
and
B.G.Vértessy
(2004).
Altered active site flexibility and a structural metal-binding site in eukaryotic dUTPase: kinetic characterization, folding, and crystallographic studies of the homotrimeric Drosophila enzyme.
|
| |
J Biol Chem,
279,
17932-17944.
|
 |
|
|
|
|
 |
L.M.Iyer,
and
L.Aravind
(2004).
The emergence of catalytic and structural diversity within the beta-clip fold.
|
| |
Proteins,
55,
977-991.
|
 |
|
|
|
|
 |
O.Barabás,
V.Pongrácz,
J.Kovári,
M.Wilmanns,
and
B.G.Vértessy
(2004).
Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase.
|
| |
J Biol Chem,
279,
42907-42915.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Mustafi,
A.Bekesi,
B.G.Vertessy,
and
M.W.Makinen
(2003).
Catalytic and structural role of the metal ion in dUTP pyrophosphatase.
|
| |
Proc Natl Acad Sci U S A,
100,
5670-5675.
|
 |
|
|
|
|
 |
E.Johansson,
O.Bjornberg,
P.O.Nyman,
and
S.Larsen
(2003).
Structure of the bifunctional dCTP deaminase-dUTPase from Methanocaldococcus jannaschii and its relation to other homotrimeric dUTPases.
|
| |
J Biol Chem,
278,
27916-27922.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Dauter,
M.Dauter,
and
E.Dodson
(2002).
Jolly SAD.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
494-506.
|
 |
|
|
|
|
 |
B.W.Han,
J.Y.Lee,
J.K.Yang,
B.I.Lee,
and
S.W.Suh
(2001).
Crystallization and preliminary X-ray crystallographic analysis of deoxyuridine triphosphate nucleotidohydrolase from Saccharomyces cerevisiae.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1147-1149.
|
 |
|
|
|
|
 |
F.Hidalgo-Zarco,
A.G.Camacho,
V.Bernier-Villamor,
J.Nord,
L.M.Ruiz-Pérez,
and
D.González-Pacanowska
(2001).
Kinetic properties and inhibition of the dimeric dUTPase-dUDPase from Leishmania major.
|
| |
Protein Sci,
10,
1426-1433.
|
 |
|
|
|
|
 |
F.Yang,
J.He,
X.Lin,
Q.Li,
D.Pan,
X.Zhang,
and
X.Xu
(2001).
Complete genome sequence of the shrimp white spot bacilliform virus.
|
| |
J Virol,
75,
11811-11820.
|
 |
|
|
|
|
 |
M.Harkiolaki,
A.M.Brzozowski,
D.Gonzalez-Pacanowska,
F.Hidalgo-Zarco,
and
K.S.Wilson
(2001).
New crystal forms of Trypanosoma cruzi dUTPase.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
915-917.
|
 |
|
|
|
|
 |
R.Persson,
M.Harkiolaki,
J.McGeehan,
and
K.S.Wilson
(2001).
Crystallization and preliminary crystallographic analysis of deoxyuridine 5'-triphosphate nucleotidohydrolase from Bacillus subtilis.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
876-878.
|
 |
|
|
|
|
 |
G.S.Prasad,
E.A.Stura,
J.H.Elder,
and
C.D.Stout
(2000).
Structures of feline immunodeficiency virus dUTP pyrophosphatase and its nucleotide complexes in three crystal forms.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
1100-1109.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Baldo,
and
M.A.McClure
(1999).
Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts.
|
| |
J Virol,
73,
7710-7721.
|
 |
|
|
|
|
 |
D.Corollo,
M.Blair-Johnson,
J.Conrad,
T.Fiedler,
D.Sun,
L.Wang,
J.Ofengand,
and
R.Fenna
(1999).
Crystallization and characterization of a fragment of pseudouridine synthase RluC from Escherichia coli.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
302-304.
|
 |
|
|
|
|
 |
V.Bernier-Villamor,
A.Camacho,
D.González-Pacanowska,
E.Cedergren-Zeppezauer,
A.Antson,
and
K.S.Wilson
(1999).
Crystallization and preliminary X-ray diffraction of Trypanosoma cruzi dUTPase.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
528-530.
|
 |
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
C.Chothia,
T.Hubbard,
S.Brenner,
H.Barns,
and
A.Murzin
(1997).
Protein folds in the all-beta and all-alpha classes.
|
| |
Annu Rev Biophys Biomol Struct,
26,
597-627.
|
 |
|
|
|
|
 |
J.M.Harris,
R.H.Haynes,
and
E.M.McIntosh
(1997).
A consensus sequence for a functional human endogenous retrovirus K (HERV-K) dUTPase.
|
| |
Biochem Cell Biol,
75,
143-151.
|
 |
|
|
|
|
 |
M.Bergdoll,
M.H.Remy,
C.Cagnon,
J.M.Masson,
and
P.Dumas
(1997).
Proline-dependent oligomerization with arm exchange.
|
| |
Structure,
5,
391-401.
|
 |
|
|
|
|
 |
R.S.Weiss,
M.O.Gold,
H.Vogel,
and
R.T.Javier
(1997).
Mutant adenovirus type 9 E4 ORF1 genes define three protein regions required for transformation of CREF cells.
|
| |
J Virol,
71,
4385-4394.
|
 |
|
|
|
|
 |
R.S.Weiss,
and
R.T.Javier
(1997).
A carboxy-terminal region required by the adenovirus type 9 E4 ORF1 oncoprotein for transformation mediates direct binding to cellular polypeptides.
|
| |
J Virol,
71,
7873-7880.
|
 |
|
|
|
|
 |
R.S.Weiss,
S.S.Lee,
B.V.Prasad,
and
R.T.Javier
(1997).
Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes.
|
| |
J Virol,
71,
1857-1870.
|
 |
|
|
|
|
 |
C.D.Mol,
J.M.Harris,
E.M.McIntosh,
and
J.A.Tainer
(1996).
Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits.
|
| |
Structure,
4,
1077-1092.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.V.Koonin
(1996).
Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases.
|
| |
Nucleic Acids Res,
24,
2411-2415.
|
 |
|
|
|
|
 |
G.Larsson,
L.A.Svensson,
and
P.O.Nyman
(1996).
Crystal structure of the Escherichia coli dUTPase in complex with a substrate analogue (dUDP).
|
| |
Nat Struct Biol,
3,
532-538.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Larsson,
P.O.Nyman,
and
J.O.Kvassman
(1996).
Kinetic characterization of dUTPase from Escherichia coli.
|
| |
J Biol Chem,
271,
24010-24016.
|
 |
|
|
|
|
 |
G.S.Prasad,
E.A.Stura,
D.E.McRee,
G.S.Laco,
C.Hasselkus-Light,
J.H.Elder,
and
C.D.Stout
(1996).
Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus.
|
| |
Protein Sci,
5,
2429-2437.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.A.Roseman,
R.K.Evans,
E.L.Mayer,
M.A.Rossi,
and
M.B.Slabaugh
(1996).
Purification and characterization of the vaccinia virus deoxyuridine triphosphatase expressed in Escherichia coli.
|
| |
J Biol Chem,
271,
23506-23511.
|
 |
|
|
|
|
 |
B.Köppe,
L.Menéndez-Arias,
and
S.Oroszlan
(1994).
Expression and purification of the mouse mammary tumor virus gag-pro transframe protein p30 and characterization of its dUTPase activity.
|
| |
J Virol,
68,
2313-2319.
|
 |
|
|
|
|
 |
E.M.McIntosh,
J.Looser,
R.H.Haynes,
and
R.E.Pearlman
(1994).
MluI site-dependent transcriptional regulation of the Candida albicans dUTPase gene.
|
| |
Curr Genet,
26,
415-421.
|
 |
|
|
|
|
 |
D.S.Threadgill,
W.K.Steagall,
M.T.Flaherty,
F.J.Fuller,
S.T.Perry,
K.E.Rushlow,
S.F.Le Grice,
and
S.L.Payne
(1993).
Characterization of equine infectious anemia virus dUTPase: growth properties of a dUTPase-deficient mutant.
|
| |
J Virol,
67,
2592-2600.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

