Exo-alpha-sialidase (GH34 Family)
The influenza virus neuramidase is a concanovin A-like lectin/glucanase. This glycoside hydrolase is composed of a central, veta-propeller catalytic domain flanked by two lectin-like domains. NA is involved in the pathogenesis of cholera by removing sialic acid from higher order gangliosides to unmask GM1, the receptor for the cholera toxin. The enzyme catalyses the hydrolysis of glycoside linkages between terminal sialic acids and adjacent sugar moieties. NA presents an excellent target for drug design against cholera.
Reference Protein and Structure
- Sequence
-
P03472
 (3.2.1.18)
(3.2.1.18)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Influenza A virus (A/tern/Australia/G70C/1975(H11N9)) (Virus)

- PDB
-
7nn9
- NATIVE INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN)
(2.0 Å)



- Catalytic CATH Domains
-
2.120.10.10
 (see all for 7nn9)
(see all for 7nn9)
Enzyme Reaction (EC:3.2.1.18)

Enzyme Mechanism
Introduction
Binding of the alpha-sialic acid to NA, forming the Michaelis complex. Triad of arginine residues binds to and stabilises the carboxylate group of the substrate by strong ionic interactions with their guanidinium groups. Distortion of the substrate pyranose ring from chair to boat conformation. The glycosidic oxygen is raised from its equatotiral position to point vertically out of the active site and lie close to an Asp 151 carboxylate oxygen. The N-acetyl group of the substrate is firmly bound in the hydrophobic pocket of the enzyme active site, with the C2 carbon forced into planar conformation. The glycerol side chain is hydrogen bonded to Glu 275 through the C8 and C9 hydroxyls. Proton donation to the glycosidic oxygen from a neighbouring water molecule, activated by a hydrogen bond to the Asp 151 residue. Glu 277 activates Tyr 412 for nucleophilic attack by an acid/base interaction between the residues. Tyr 412 is close to, and nucleophilically attacks the anomeric carbon centre of the sialic acid residue. Formation and stabilisation of the endocyclic sialosyl cation transition state. Formation and release of the sialic acid as the alpha-monomer
Catalytic Residues Roles
| UniProt | PDB* (7nn9) | ||
| Asp152 | Asp151(70)A | Acts as a general acid catalyst. The hydroxyl group of sialic acid at C4 interacts with OD2 of Asp 151. Asp 151 stabilises the proton-donating water molecule via hydrogen bonding. | proton acceptor, proton donor, activator, electrostatic stabiliser, increase acidity, increase nucleophilicity |
| Glu279 | Glu277(197)A | Acts as a general base that facilitates nucleophilic attack of the anomeric carbon by Tyr 412. The hydroxyls OH8 and OH9 of the glycerol side chain of sialic acid are hydrogen bonded to Glu 277. This stabilises the sialosyl cation intermediate. Interaction with Arg 220 stabilises the Arg position within the arginine triad. | proton acceptor, proton donor, activator, electrostatic stabiliser, increase nucleophilicity, promote heterolysis |
| Tyr406 | Tyr406(324)A | Tyr 412 is positioned close to, and nucleophilically attacks the anomeric carbon of sialic acid. This is mediated by an acid/base interaction with Glu 277. | nucleofuge, nucleophile, proton acceptor, proton donor, electrostatic stabiliser |
| Arg372, Arg294 | Arg371(290)A, Arg292(212)A | Catalytic since the carboxylate oxygens, O1 and O2, have close contact to N1 and N2 of the residue. Therefore, this residue is involved in stabilisation of the carboxylate. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, intermediate formation, overall product formed, native state of enzyme regenerated, hydrolysis, intermediate terminatedReferences
- Vavricka CJ et al. (2013), Nat Commun, 4, 1491-. Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors. DOI:10.1038/ncomms2487. PMID:23422659.
- Hinou H et al. (2005), Biochemistry, 44, 11669-11675. Characterization ofVibrio choleraeNeuraminidase by a Novel Mechanism-Based Fluorescent Labeling Reagent. DOI:10.1021/bi0509954. PMID:16128567.
- Buschiazzo A et al. (2000), EMBO J, 19, 16-24. Structural basis of sialyltransferase activity in trypanosomal sialidases. DOI:10.1093/emboj/19.1.16. PMID:10619840.
- Ghate AA et al. (1998), Eur J Biochem, 258, 320-331. Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design. DOI:10.1046/j.1432-1327.1998.2580320.x. PMID:9874196.
- Davies G et al. (1995), Structure, 3, 853-859. Structures and mechanisms of glycosyl hydrolases. DOI:10.1016/s0969-2126(01)00220-9. PMID:8535779.

Step 1. Glu277 activates Tyr412 which attacks C1. The sialyl ketal linkage is broken in an SN2 reaction with Asp151 protonating a water molecule which in turn protonates the leaving group the leaving group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu277(197)A | electrostatic stabiliser |
| Asp151(70)A | electrostatic stabiliser |
| Arg292(212)A | electrostatic stabiliser |
| Tyr406(324)A | electrostatic stabiliser |
| Arg371(290)A | electrostatic stabiliser |
| Glu277(197)A | activator, increase nucleophilicity |
| Asp151(70)A | increase acidity, proton donor |
| Glu277(197)A | proton acceptor |
| Tyr406(324)A | proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, intermediate formation, overall product formed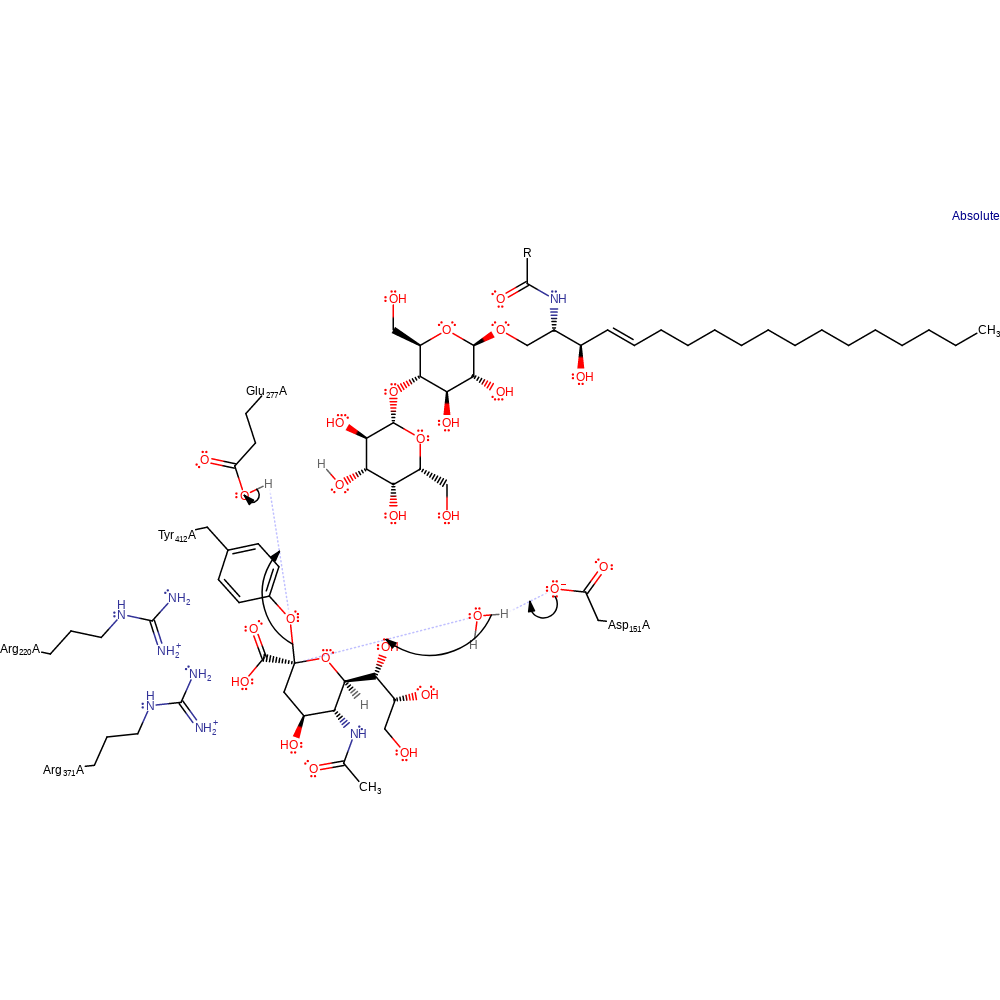
Step 2. Asp92 activates the water molecule which hydrolyses the tyrosine bound intermediate. Glu277 protonates the departing Tyr412.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp151(70)A | electrostatic stabiliser |
| Arg292(212)A | electrostatic stabiliser |
| Glu277(197)A | electrostatic stabiliser |
| Arg371(290)A | electrostatic stabiliser |
| Asp151(70)A | activator, increase nucleophilicity |
| Glu277(197)A | promote heterolysis |
| Asp151(70)A | proton acceptor |
| Tyr406(324)A | proton acceptor |
| Glu277(197)A | proton donor |
| Tyr406(324)A | nucleofuge |



 Download:
Download: