Uridine phosphorylase
The enzyme uridine phosphorylase is very important in the pathway whereby uracil can be scavenged to increase blood uracil levels. The overall reaction thus results in the formation of ribose-1-phosphate in addition to uracil. The pathway is of particular interest because it provides a mechanism whereby the effect of base analogues such as 5BU can be minimised, thus may provide some defence against tumour formation. The enzyme itself shows significant homology to Purine nucleotide phosphorylases rather than to pyrimidine nucleotide phosphorylases, posing interesting questions about its evolution.
Reference Protein and Structure
- Sequence
-
P12758
 (2.4.2.3)
(2.4.2.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1t0u
- Crystal structure of E.coli uridine phosphorylase at 2.2 A resolution (Type-A Native)
(2.2 Å)



- Catalytic CATH Domains
-
3.40.50.1580
 (see all for 1t0u)
(see all for 1t0u)
Enzyme Reaction (EC:2.4.2.3)
Enzyme Mechanism
Introduction
The reaction mechanism is believed to be similar to that of human purine nucleotide phosphorylase, so involves an carboxenium ion transition state formed through electron density from the 4' oxygen being pushed onto the 1' Carbon of the ribosyl moiety. The formation of this transition state is assisted by transient deprotonation of the 5' OH group by His 8, primed by Glu 80, which is positioned directly above the ribosyl ring oxygen by the binding of the substrate in the active site. The release of electrons towards the 1' Carbon facilitates the cleavage of the CN bond to release uracil. Protonation of the uracil moiety prior to the cleavage of the bond occurs through a water molecule activated by Arg 223, and this allows it to act as a leaving group whilst the O4 atom of the phosphate acts as a nucleophile to attack the 1' Carbon. As a result, the products are formed.
Catalytic Residues Roles
| UniProt | PDB* (1t0u) | ||
| His8 | His8A | Acts as a general base to deprotonate the 5'OH to allow the build up of negative charge that results in the formation of the oxocarbenium ion transition state. | proton acceptor, proton donor |
| Glu80 | Glu80A | Acts to modify the pKa of His 8 to allow it to act as a general acid base and deprotonate the 5'OH group of the ribosyl moiety, thus allowing formation of the oxocarbenium ion transition state. | electrostatic stabiliser, polar interaction |
| Arg223 | Arg223B | Acts to allow a water molecule to act as an acid base and protonate the uracil moiety, thus allowing it to act as a leaving group. | electrostatic stabiliser, polar interaction |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, heterolysis, inferred reaction step, native state of enzyme regeneratedReferences
- Caradoc-Davies TT et al. (2004), J Mol Biol, 337, 337-354. Crystal Structures of Escherichia coli Uridine Phosphorylase in Two Native and Three Complexed Forms Reveal Basis of Substrate Specificity, Induced Conformational Changes and Influence of Potassium. DOI:10.1016/j.jmb.2004.01.039. PMID:15003451.
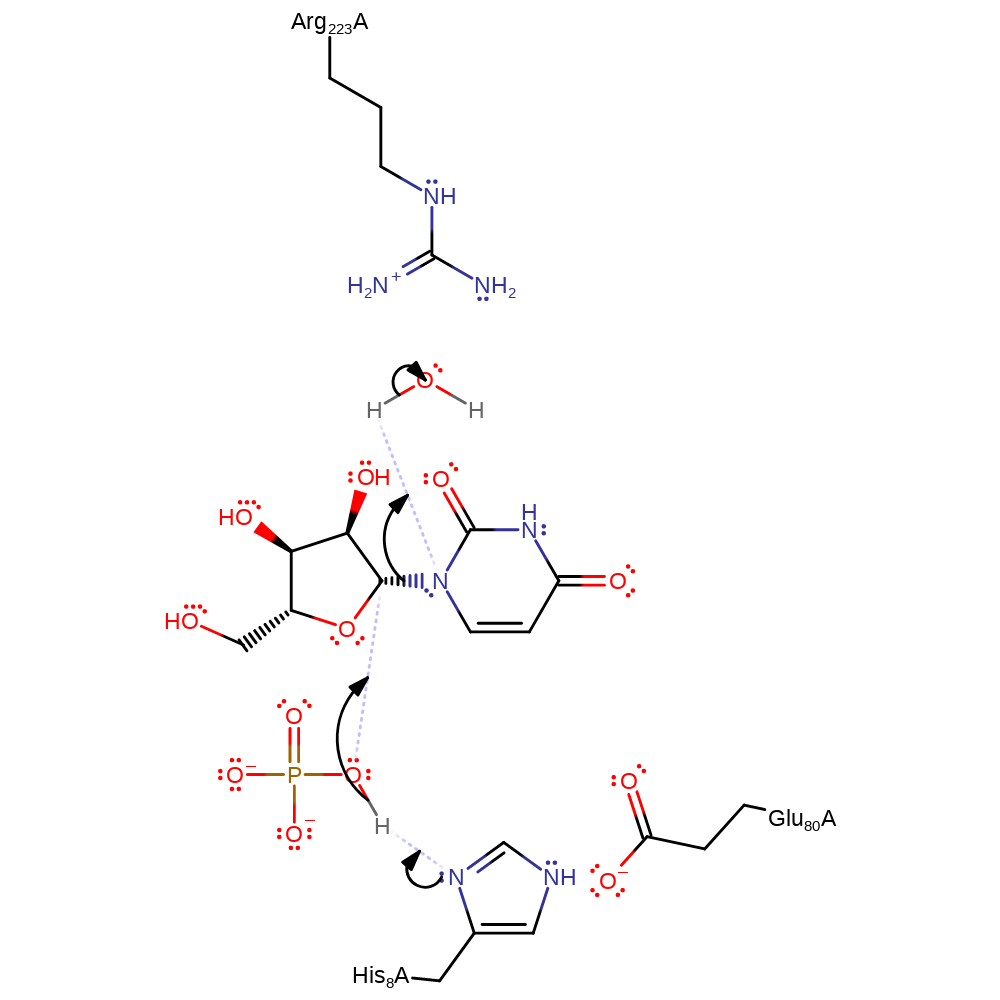
Step 1. Arg 223 forms hydrogen bonds with the proton donating water that donates a proton to the uracil base. This His8 deprotonates O5' on the phosphate group, making it more electron rich and facilitating nucleophilic attack.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu80A | electrostatic stabiliser |
| Arg223B | electrostatic stabiliser |
| Glu80A | polar interaction |
| Arg223B | polar interaction |
| His8A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, heterolysis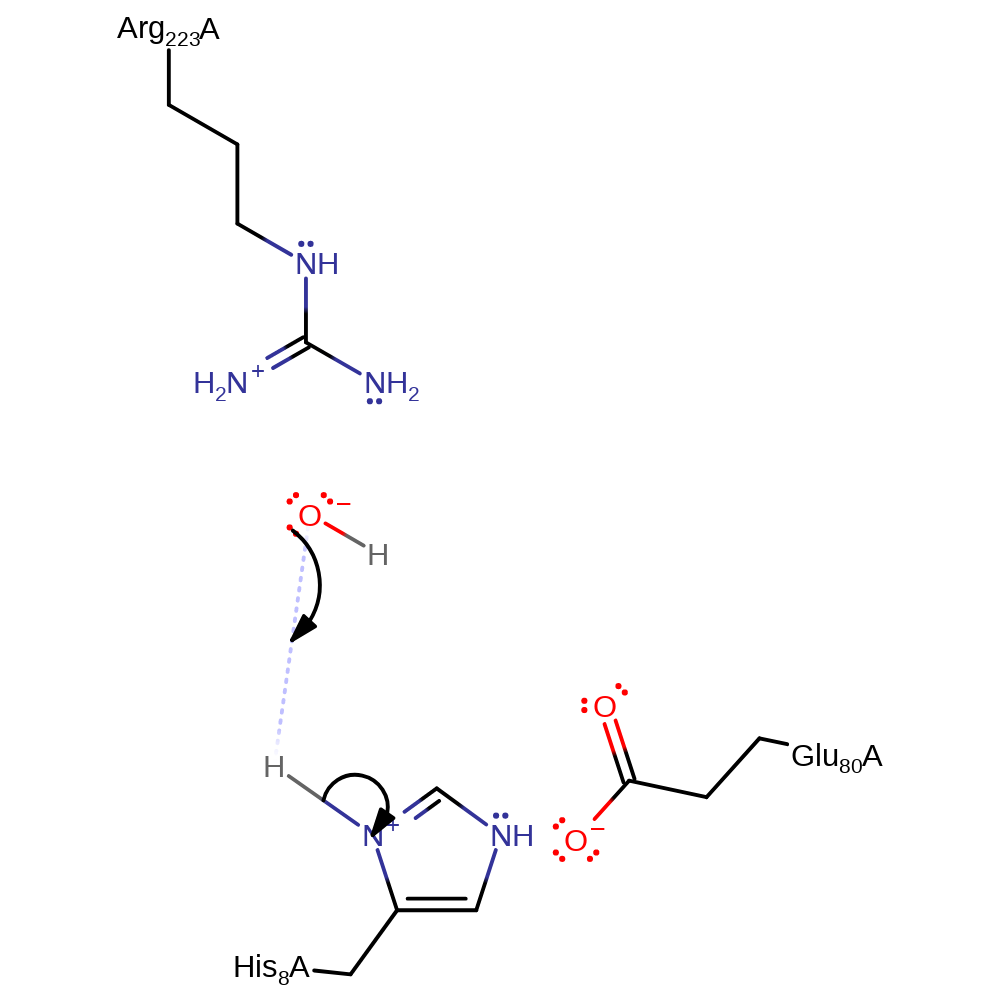
Step 2. His8 is deprotonated by the hydroxide ion, ready for another round of catalysis.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His8A | proton donor |
Chemical Components
inferred reaction step, proton transfer, native state of enzyme regeneratedIntroduction
Uracil in the active site is in a strained C4'- endo conformation. This promotes glycosidic bond cleavage in the strained N-C1' bond. Subsequently Arg168 protonates the developing negatively charged uracil base while an oxygen nucleophile on the phosphate attacks the oxycarbenium ion to form ribose 1-phosphate in an Sn1 reaction. This mechanism is favoured when compared to proton donation from a water hydrogen bonded to Arg223 due to mutagenesis experiments demonstrating Arg168 being a critical residue for enzymatic activity.
Catalytic Residues Roles
| UniProt | PDB* (1t0u) | ||
| Arg168 | ArgNone(168)A | Stabilises the developing negative charge on the uracil by protonating the base. | proton acceptor, proton donor |
Chemical Components
proton transfer, overall reactant used, heterolysis, elimination (not covered by the Ingold mechanisms), intermediate formation, intermediate terminated, bimolecular nucleophilic addition, inferred reaction step, native state of enzyme regenerated, overall product formedReferences
- Bu W et al. (2005), Acta Crystallogr D Biol Crystallogr, 61, 863-872. Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines. DOI:10.1107/S0907444905007882. PMID:15983408.
- Lashkov AA et al. (2010), 55, 41-57. Structural basis for the mechanism of inhibition of uridine phosphorylase from Salmonella typhimurium. DOI:10.1134/S1063774510010098.
- Caradoc-Davies TT et al. (2004), J Mol Biol, 337, 337-354. Crystal Structures of Escherichia coli Uridine Phosphorylase in Two Native and Three Complexed Forms Reveal Basis of Substrate Specificity, Induced Conformational Changes and Influence of Potassium. DOI:10.1016/j.jmb.2004.01.039. PMID:15003451.
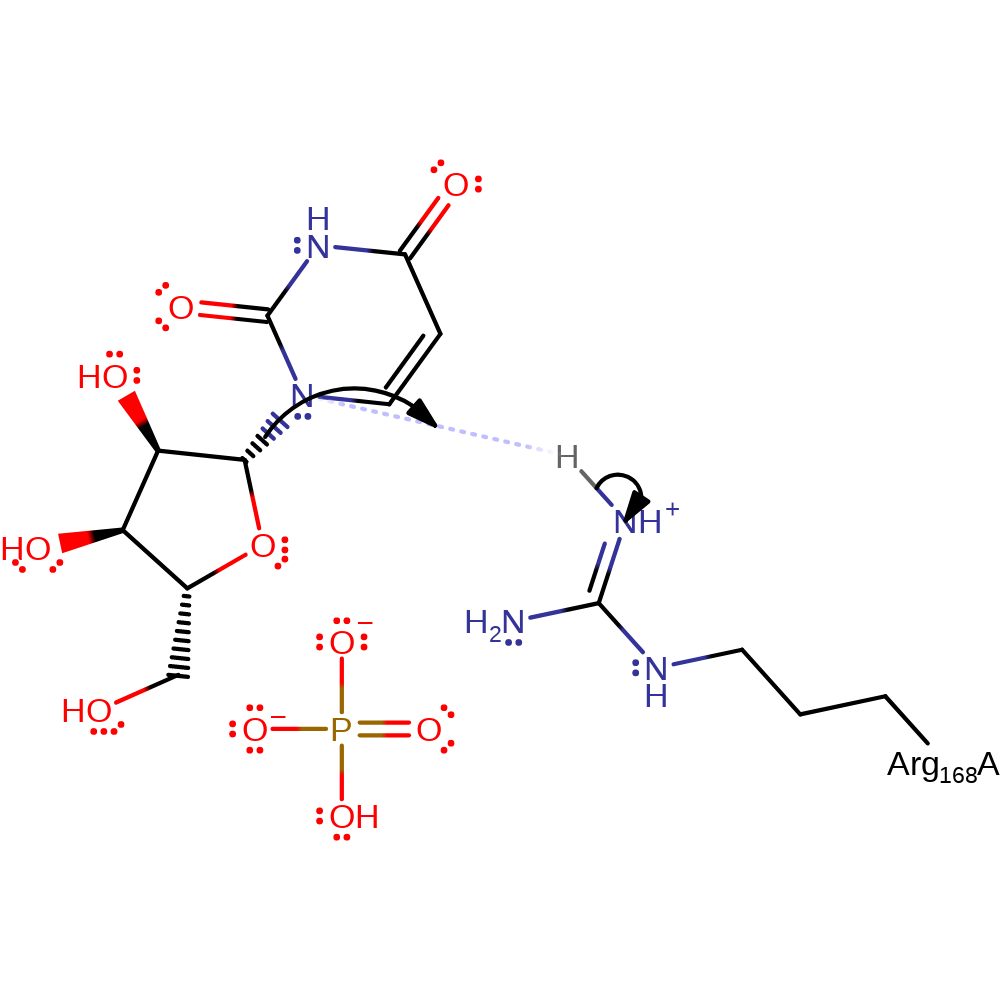
Step 1. Substrate uridine being in a strained state upon binding to the enzyme results in glycosidic bond cleavage. Negatively charged uracil is protonated by Arg168.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| ArgNone(168)A | proton donor |
Chemical Components
proton transfer, overall reactant used, heterolysis, elimination (not covered by the Ingold mechanisms), intermediate formation
Step 2. Nucleophilic attack on the C1 oxocarbenium ion by O on phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
intermediate terminated, ingold: bimolecular nucleophilic addition, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| ArgNone(168)A | proton acceptor |
Chemical Components
proton transfer, inferred reaction step, native state of enzyme regenerated, overall product formedIntroduction
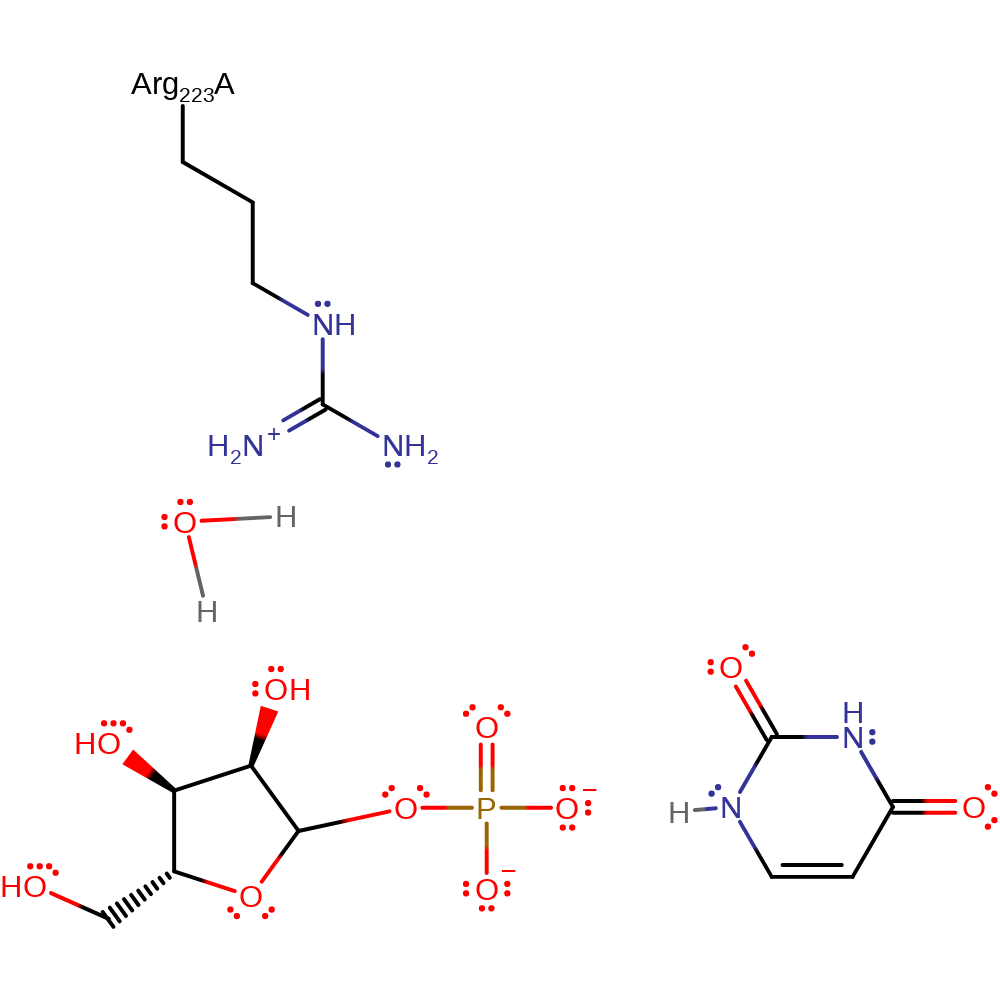
As with mechanism proposal 2, uridine is in a strained C4'-endo conformation which promotes glycosidic bond cleavage. The developing negative charges of the base are stabilised by proton transfer from a water molecule hydrogen bonded to Arg223 while the resulting oxocarbenium ion transition state collapses on attack from an O atom on the phosphate group. The final reaction products are ribose-1-phosphate and uracil.
Catalytic Residues Roles
| UniProt | PDB* (1t0u) | ||
| Arg223 | Arg223B | Hydrogen bonds to a water molecule that a donates a proton to the leaving group uracil at N7. | electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, heterolysis, intermediate formation, bimolecular nucleophilic addition, intermediate terminated, overall product formed, inferred reaction stepReferences
- Bu W et al. (2005), Acta Crystallogr D Biol Crystallogr, 61, 863-872. Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines. DOI:10.1107/S0907444905007882. PMID:15983408.
- Lashkov AA et al. (2010), 55, 41-57. Structural basis for the mechanism of inhibition of uridine phosphorylase from Salmonella typhimurium. DOI:10.1134/S1063774510010098.
- Caradoc-Davies TT et al. (2004), J Mol Biol, 337, 337-354. Crystal Structures of Escherichia coli Uridine Phosphorylase in Two Native and Three Complexed Forms Reveal Basis of Substrate Specificity, Induced Conformational Changes and Influence of Potassium. DOI:10.1016/j.jmb.2004.01.039. PMID:15003451.

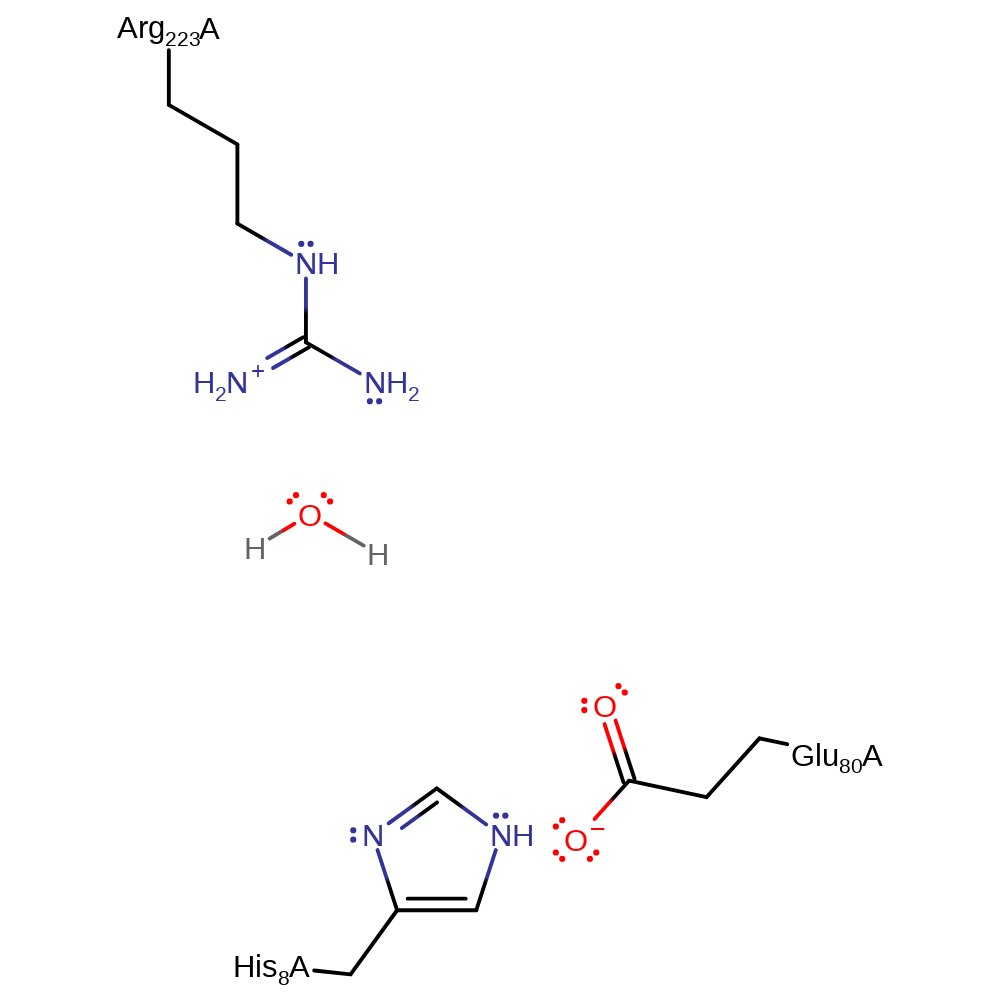
Step 1. Strained uridine promotes glycosidic bond cleavage. The developing negatively charged base is stabilised by protonation by a water molecule hydrogen bonded to Arg223.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg223B | electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, heterolysis, intermediate formation
Step 2. The oxocarbenium ion is subject to nucleophilic attack of an O atom of the phosphate group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate terminated, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|



 Download:
Download: 
 Download:
Download: 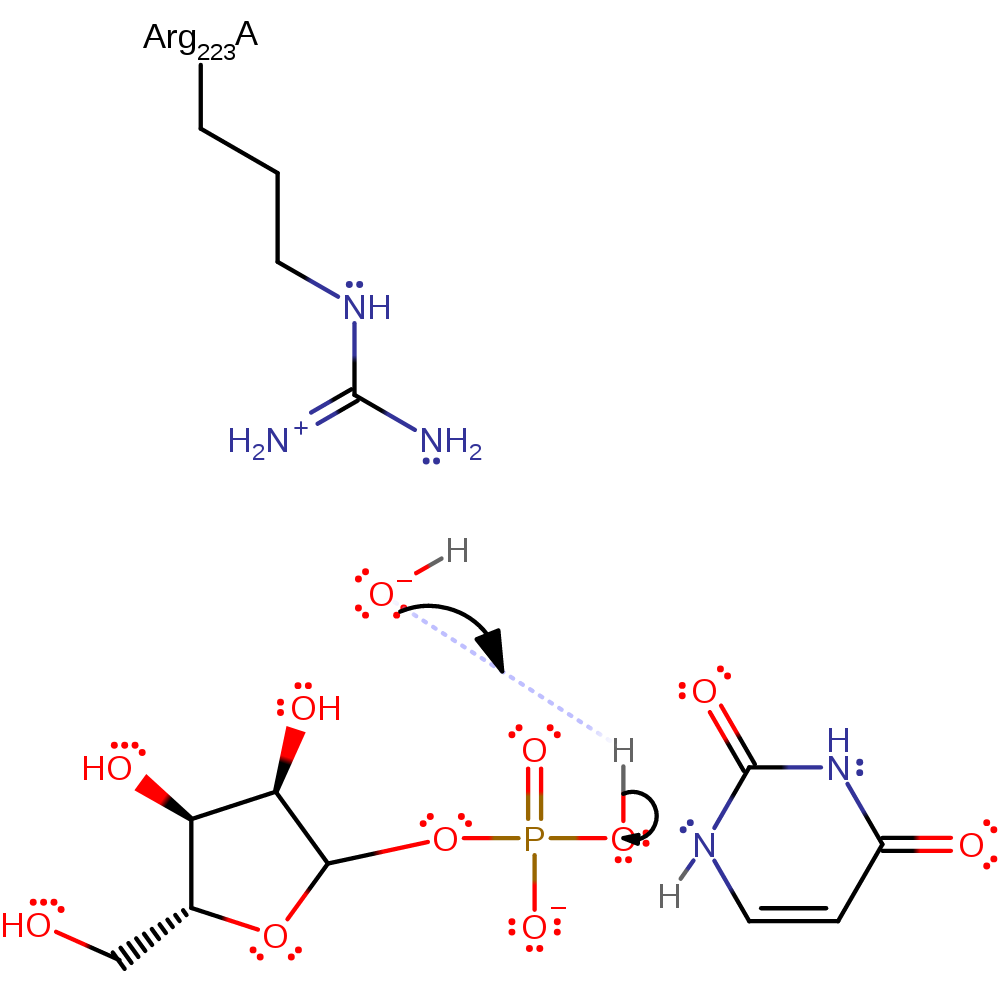
 Download:
Download: