Myristoyl-ACP-specific thioesterase
Myristoyl acyl carrier protein specific thioesterase (C14ACP-TE) is a lux-specific acyltransferase belonging to the alpha/beta hydrolase family. The acyl transferase is part of the fatty acid reductase system required for aldehyde biosynthesis. It produces fatty acids for the light-emitting reaction catalysed by luciferase in luminescent bacteria. In Vibrio harveyi, C14ACP-TE is responsible for catalysing the transfer or hydrolysis of acyl group from either myristoyl-ACP or myristoyl-CoA to form myristic acid, and for diverting the myristic acid to the bioluminescence pathway, where it undergoes NADPH-dependent reduction and subsequent FMNH2- and O2-dependent oxidation of the corresponding aldehyde, with accompanying emission of light.
Reference Protein and Structure
- Sequence
-
P05521
 (2.3.1.-)
(2.3.1.-)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Vibrio harveyi (Bacteria)

- PDB
-
1tht
- STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI
(2.1 Å)



- Catalytic CATH Domains
-
3.40.50.1820
 (see all for 1tht)
(see all for 1tht)
Enzyme Mechanism
Introduction
Enzymatic hydrolysis of the thioester bond proceeds using the catalytic triad of Ser 114, Asp 211, and His 241. His 241 activates Ser 114 as a nucleophile by abstracting a proton from the serine hydroxyl as the carbonyl carbon of the substrate is attacked by the nucleophile. His 241 is stabilised by hydrogen bonding to Asp 211 which lowers the pKa of His so that it more willingly accepts a proton. This results in the formation of a tetrahedral transition state which collapses, with the His 241 donating a proton to the first leaving group (CoA), to form the acylenzyme intermediate. The intermediate is subsequently attacked by the activated ACP sulphur as it is deprotonated by His 241 to activate it. This second tetrahedral transition state will collapse resulting in the cleavage of the acyl-enzyme bond and the released Ser 114 will accept a proton from His 241 which regenerates the native state of the active site.
Catalytic Residues Roles
| UniProt | PDB* (1tht) | ||
| Asp211 | Asp211A | Stabilises His 241 by hydrogen bonding which as a result lowers His 241's pKa and so will more willingly accept a proton from Ser 114. | modifies pKa, electrostatic stabiliser |
| Ser114 | Ser114A | Ser114 is activated by His241. It acts as a nucleophile and attacks the substrate to form the acylenzyme intermediate. Its backbone amide may also be involved in the formation of an oxyanion hole. | covalently attached, nucleofuge, nucleophile, proton acceptor, proton donor |
| His241 | His241A | His241 acts as a general acid/base catalyst, activating Ser114 and water as nucleophiles. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, enzyme-substrate complex cleavage, native state of enzyme regeneratedReferences
- Li J et al. (1996), Biochemistry, 35, 9967-9973. Conversion of Serine-114 to Cysteine-114 and the Role of the Active Site Nucleophile in Acyl Transfer by Myristoyl-ACP Thioesterase fromVibrio harveyi†. DOI:10.1021/bi9605292. PMID:8756458.
- Lawson DM et al. (1994), Biochemistry, 33, 9382-9388. Structure of a Myristoyl-ACP-Specific Thioesterase from Vibrio harveyi. DOI:10.1021/bi00198a003. PMID:8068614.
- Ferri SR et al. (1994), J Biol Chem, 269, 6683-6688. An essential histidine residue required for fatty acylation and acyl transfer by myristoyltransferase from luminescent bacteria. PMID:8120025.

Step 1. His241 abstracts a proton from Ser114 and activates it for nucleophilic attack on the thioester carbonyl of myristoyl-CoA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser114A | covalently attached |
| Asp211A | electrostatic stabiliser, modifies pKa |
| His241A | proton acceptor |
| Ser114A | proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used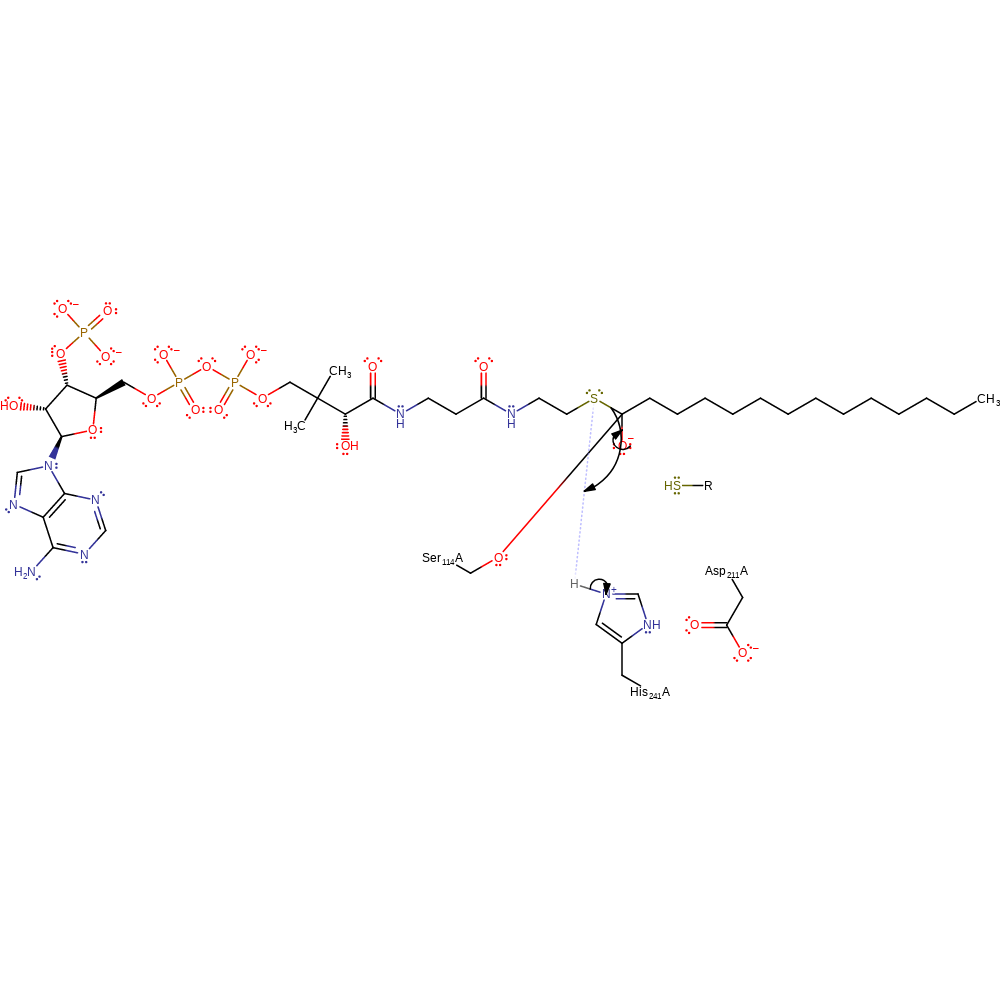
Step 2. His241 protonates the sulphur that bridges the CoA extremity and myristoyl which results in the collapse of the tetrahedral intermediate and the cleavage of the S-C bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp211A | electrostatic stabiliser, modifies pKa |
| Ser114A | covalently attached |
| His241A | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, intermediate collapse, overall product formed
Step 3. His241 deprotonates the the sulphur of ACP which activates it to nucleophilically attack the carbonyl carbon of the acyl-enzyme intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp211A | electrostatic stabiliser, modifies pKa |
| Ser114A | covalently attached |
| His241A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 4. The tetrahedral intermediate collapses resulting in the cleavage of the acyl-enzyme bond which releases Ser 114 which can now accept a proton from His 241.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp211A | electrostatic stabiliser, modifies pKa |
| Ser114A | covalently attached |
| His241A | proton donor |
| Ser114A | nucleofuge, proton acceptor |




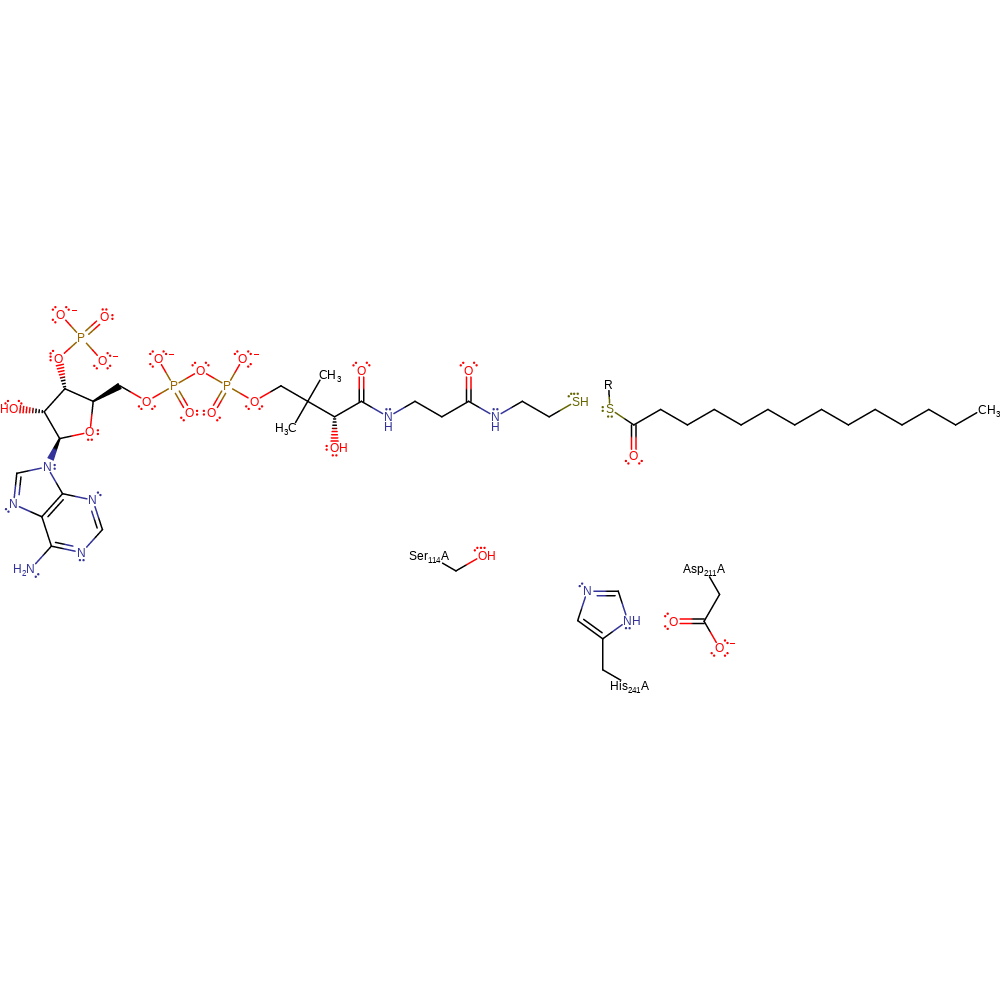 Download:
Download: