Aldehyde reductase
This entry represents the aldo-keto reductase family which includes a number of related monomeric NAD(P)H-dependent oxidoreductases, such as aldehyde reductase, aldose reductase, prostaglandin F synthase, xylose reductase, rho crystallin, etc. [PMID:2498333].
Aldehyde Reductase, sourced from Homo sapiens, catalyses the NADPH-dependent reduction of a wide range of carbonyl compounds such as sugars, aldehyde metabolites, corticosteroid hormones and xenobiotic aldehydes. Interest in this enzyme stems from it's ability to reduce glucose and galactose, causing diabetic and galactosemic complications affecting the lens, retina, nerves and kidneys.
Reference Protein and Structure
- Sequence
-
P15121
 (1.1.1.21, 1.1.1.54, 1.1.1.300, 1.1.1.372)
(1.1.1.21, 1.1.1.54, 1.1.1.300, 1.1.1.372)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
2acu
- TYROSINE-48 IS THE PROTON DONOR AND HISTIDINE-110 DIRECTS SUBSTRATE STEREOCHEMICAL SELECTIVITY IN THE REDUCTION REACTION OF HUMAN ALDOSE REDUCTASE: ENZYME KINETICS AND THE CRYSTAL STRUCTURE OF THE Y48H MUTANT ENZYME
(1.76 Å)



- Catalytic CATH Domains
-
3.20.20.100
 (see all for 2acu)
(see all for 2acu)
Enzyme Reaction (EC:1.1.1.2)
Enzyme Mechanism
Introduction
The pro-R hydrogen of NAD(P)H is transferred to the re face of the carbonyl group of the substrate. The forming alcohol group is protonated by Tyr 48, which acts as a general acid. The Tyr 48 anion is stabilised electrostatically by Lys 77. Lys 77 is stabilised by Asp 43.
Catalytic Residues Roles
| UniProt | PDB* (2acu) | ||
| Tyr49 | His48A | The forming alcohol group is protonated by Tyr 48, which acts as a general acid. This function is facilitated by a hydrogen bonding network with Lys77 and Asp43. | proton acceptor, proton donor |
| His111 | His110A | This histidine plays a major role in determining the substrate and stereo- specificity of these enzymes. | electrostatic stabiliser |
| Asp44 | Asp43A | Asp 43 electrostatically stabilises Lys 77. | electrostatic stabiliser |
| Lys78 | Lys77A | Lys 77 stabilises the Tyr 48 anion. | modifies pKa, electrostatic stabiliser |
Chemical Components
proton transfer, hydride transfer, overall reactant used, overall product formed, inferred reaction step, native state of enzyme regeneratedReferences
- Kratzer R et al. (2006), IUBMB Life, 58, 499-507. Catalytic mechanism and substrate selectivity of aldo-keto reductases: insights from structure-function studies of Candida tenuis xylose reductase. DOI:10.1080/15216540600818143. PMID:17002977.
- Liu X et al. (2014), Protein Sci, 23, 1540-1549. Structural and mutational studies on an aldo-keto reductase AKR5C3 from Gluconobacter oxydans. DOI:10.1002/pro.2531. PMID:25131535.
- Chen LC et al. (2009), Acta Crystallogr Sect F Struct Biol Cryst Commun, 65, 419-421. Purification, crystallization and preliminary X-ray crystallographic analysis of xylose reductase from Candida tropicalis. DOI:10.1107/S1744309109008719. PMID:19342796.
- Olsen JG et al. (2008), Proteins, 71, 1572-1581. Barley aldose reductase: structure, cofactor binding, and substrate recognition in the aldo/keto reductase 4C family. DOI:10.1002/prot.21996. PMID:18300247.
- Bains OS et al. (2008), Drug Metab Dispos, 36, 904-910. Two allelic variants of aldo-keto reductase 1A1 exhibit reduced in vitro metabolism of daunorubicin. DOI:10.1124/dmd.107.018895. PMID:18276838.
- Pival SL et al. (2008), FEBS Lett, 582, 4095-4099. Tyr-51 is the proton donor-acceptor for NAD(H)-dependent interconversion of xylose and xylitol by Candida tenuis xylose reductase (AKR2B5). DOI:10.1016/j.febslet.2008.11.003. PMID:19026644.
- Kratzer R et al. (2005), Biochem J, 389, 507-515. Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida tenuis xylose reductase (AKR2B5) probed by site-directed mutagenesis and functional complementation studies. DOI:10.1042/BJ20050167. PMID:15799715.
- El-Kabbani O et al. (2005), J Med Chem, 48, 5536-5542. Structure of aldehyde reductase holoenzyme in complex with the potent aldose reductase inhibitor fidarestat: implications for inhibitor binding and selectivity. DOI:10.1021/jm050412o. PMID:16107153.
- Bohren KM et al. (2005), Biochim Biophys Acta, 1748, 201-212. The structure of Apo R268A human aldose reductase: hinges and latches that control the kinetic mechanism. DOI:10.1016/j.bbapap.2005.01.006. PMID:15769597.
- Ruiz F et al. (2004), Acta Crystallogr D Biol Crystallogr, 60, 1347-1354. The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48. DOI:10.1107/S0907444904011370. PMID:15272156.
- Sanli G et al. (2004), Protein Sci, 13, 504-512. Structural alteration of cofactor specificity in Corynebacterium 2,5-diketo-D-gluconic acid reductase. DOI:10.1110/ps.03450704. PMID:14718658.
- Kratzer R et al. (2004), Biochemistry, 43, 4944-4954. Studies of the enzymic mechanism of Candida tenuis xylose reductase (AKR 2B5): X-ray structure and catalytic reaction profile for the H113A mutant. DOI:10.1021/bi035833r. PMID:15109252.
- Vander Jagt DL et al. (2003), ACS Symp Ser Am Chem Soc, 23-35. Aldo-Keto Reductase-Catalyzed Detoxication of Endogenous Aldehydes Associated with Diabetic Complications. DOI:10.1021/bk-2003-0865.ch002.
- Palackal NT et al. (2001), Chem Biol Interact, 130-132, 815-824. Metabolic activation of polycyclic aromatic hydrocarbon trans-dihydrodiols by ubiquitously expressed aldehyde reductase (AKR1A1). PMID:11306097.
- Ye Q et al. (2000), Proteins, 38, 41-48. Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily. PMID:10651037.
- Várnai P et al. (1999), Proteins, 37, 218-227. Modelling the catalytic reaction in human aldose reductase. PMID:10584067.
- O'connor T et al. (1999), Biochem J, 343 Pt 2, 487-504. Major differences exist in the function and tissue-specific expression of human aflatoxin B1 aldehyde reductase and the principal human aldo-keto reductase AKR1 family members. PMID:10510318.
- Khurana S et al. (1998), Proc Natl Acad Sci U S A, 95, 6768-6773. Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution. DOI:10.1073/pnas.95.12.6768. PMID:9618487.
- Jez JM et al. (1997), Biochem J, 326, 625-636. Comparative anatomy of the aldo–keto reductase superfamily. DOI:10.1042/bj3260625. PMID:9307009.
- el-Kabbani O et al. (1995), Nat Struct Biol, 2, 687-692. Structure of porcine aldehyde reductase holoenzyme. DOI:10.1038/nsb0895-687. PMID:7552731.
- Barski OA et al. (1995), Biochemistry, 34, 11264-11275. Mechanism of human aldehyde reductase: characterization of the active site pocket. PMID:7669785.
- Bohren KM et al. (1994), Biochemistry, 33, 2021-2032. Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme. DOI:10.2210/pdb2acu/pdb. PMID:8117659.
- El-Kabbani O et al. (1994), Acta Crystallogr D Biol Crystallogr, 50, 859-868. Structures of human and porcine aldehyde reductase: an enzyme implicated in diabetic complications. DOI:10.1107/S0907444994005275. PMID:15299353.
- Bruce NC et al. (1994), Biochem J, 299, 805-811. Bacterial morphine dehydrogenase further defines a distinct superfamily of oxidoreductases with diverse functional activities. DOI:10.1042/bj2990805. PMID:8192670.
- Tarle I et al. (1993), J Biol Chem, 268, 25687-25693. Probing the active site of human aldose reductase. Site-directed mutagenesis of Asp-43, Tyr-48, Lys-77, and His-110. PMID:8245005.
- Wilson DK et al. (1992), Science, 257, 81-84. An unlikely sugar substrate site in the 1.65 A structure of the human aldose reductase holoenzyme implicated in diabetic complications. DOI:10.1126/science.1621098. PMID:1621098.
- Bohren KM et al. (1989), J Biol Chem, 264, 9547-9551. The aldo-keto reductase superfamily. cDNAs and deduced amino acid sequences of human aldehyde and aldose reductases. PMID:2498333.
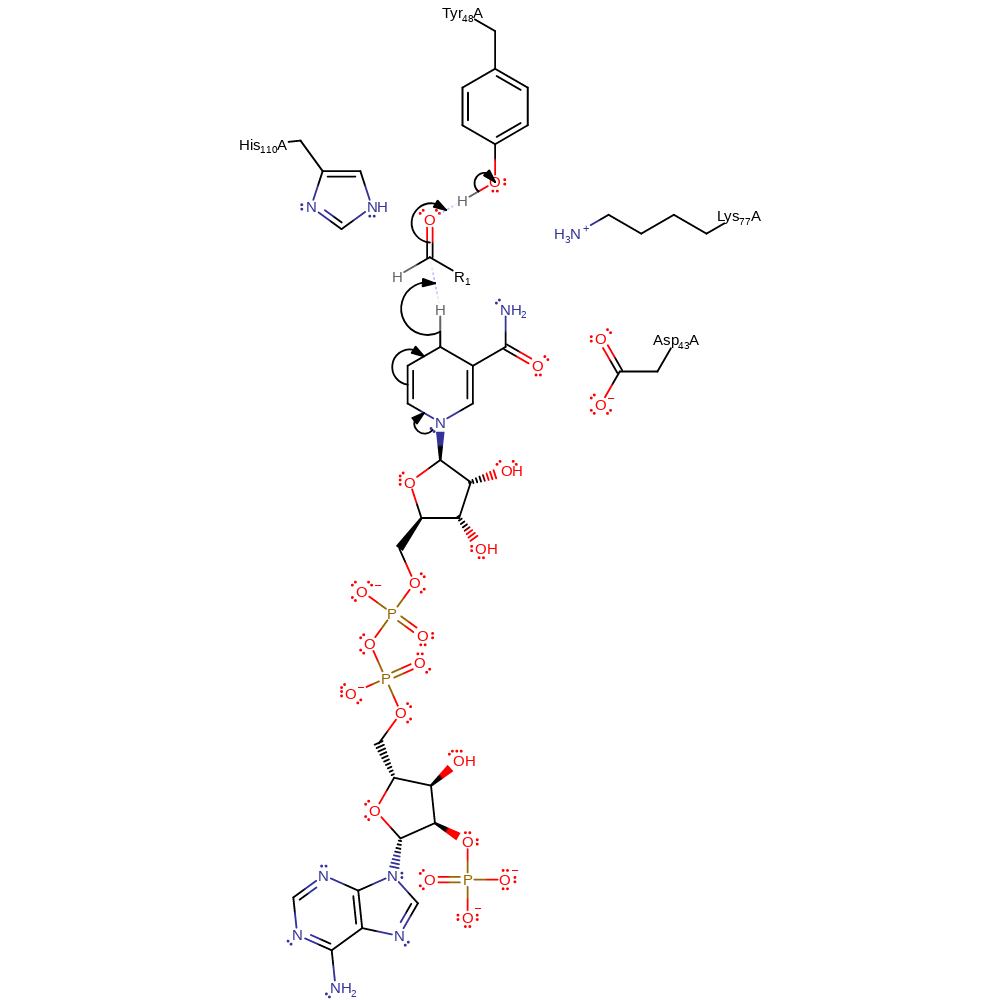
Step 1. NAD(P)H eliminated a hydride from the pro-R hydrogen to the pseudo re-side of the ketone. Re- and si-side are denoted according to the relative steric requirements of the carbonyl group substituents where the H represents a small group and R1 the large group. The ketone then abstracts a proton from the tyrosine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp43A | electrostatic stabiliser |
| Lys77A | electrostatic stabiliser, modifies pKa |
| His110A | electrostatic stabiliser |
| His48A | proton donor |
Chemical Components
proton transfer, hydride transfer, overall reactant used, overall product formed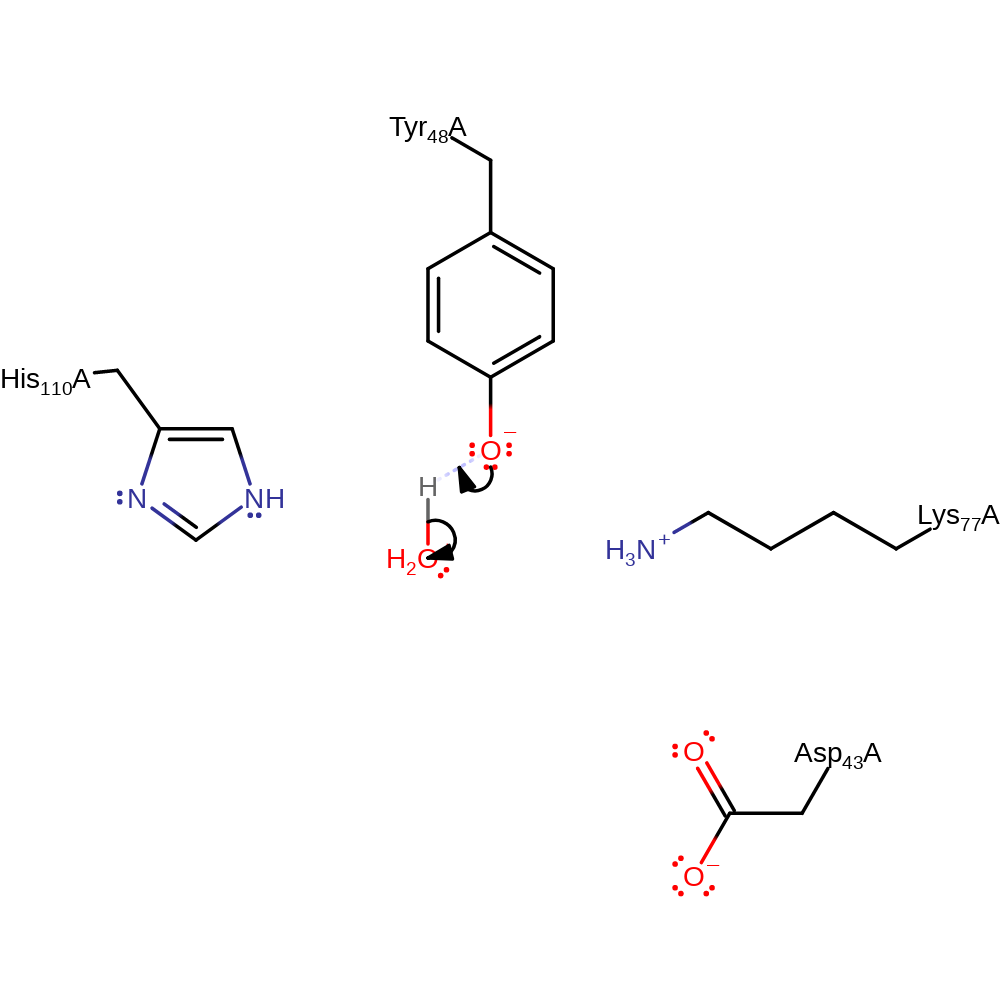
Step 2. In an inferred return step, water donates a proton to the active site tyrosine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp43A | electrostatic stabiliser |
| Lys77A | modifies pKa, electrostatic stabiliser |
| His48A | proton acceptor |





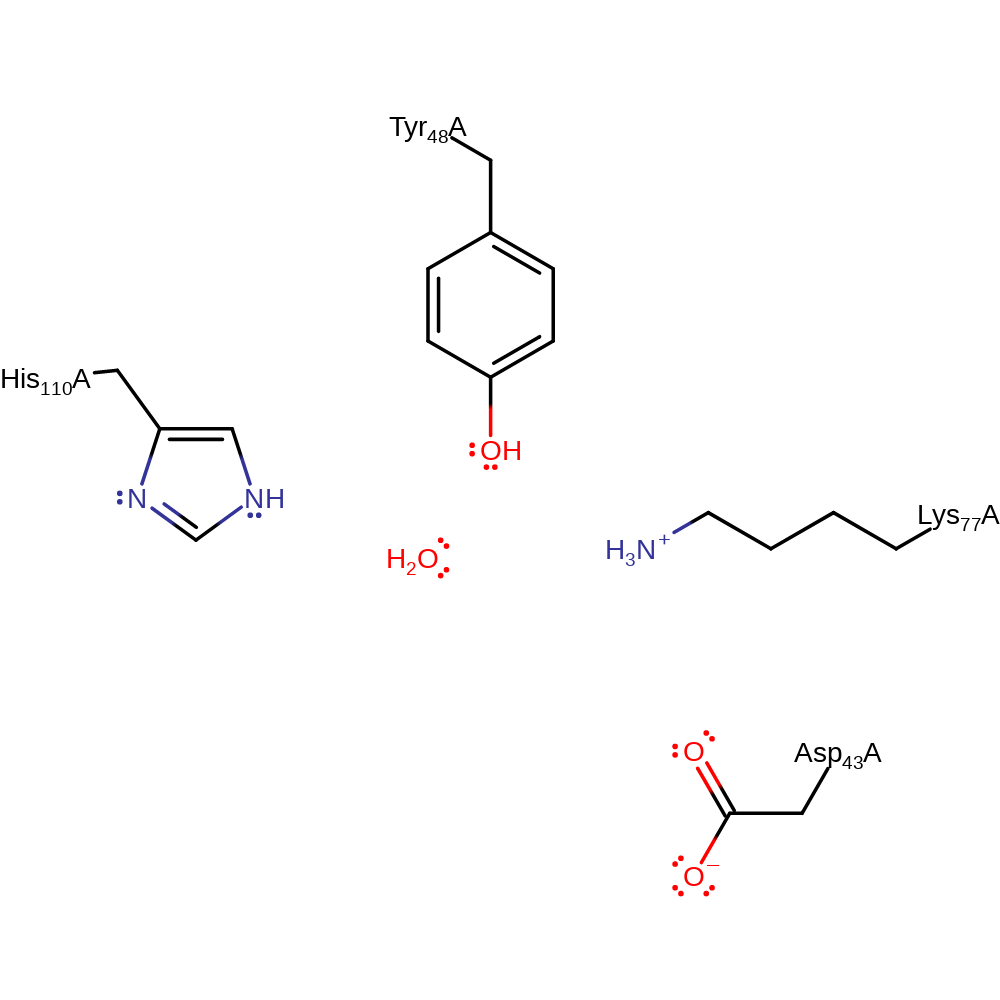 Download:
Download: