Rhamnogalacturonan acetylesterase
Rhamnogalacturonan acetyltransferase (RGAE) is one of a group of enzymes produced by Aspergillus aculeatus to perform the synergistic degradation of rhamnogalacturonan, a complex polysaccharide found in pectic substances that constitute part of the middle lamella and the primary cell wall of higher plants. RGAE catalyses the deacylation of the backbone of rhamnogalacturonan, which is essential for the subsequent action of enzymes that cleave the glycosidic bonds and so degrade the polysaccharide. RGAE is a member of the carbohydrate esterase family 12, performing hydrolysis on an ester bond using a Ser-His-Asp catalytic triad.
Reference Protein and Structure
- Sequence
-
Q00017
 (3.1.1.86)
(3.1.1.86)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Aspergillus aculeatus (Fungus)

- PDB
-
1pp4
- The crystal structure of rhamnogalacturonan acetylesterase in space group P3121
(2.5 Å)



- Catalytic CATH Domains
-
3.40.50.1110
 (see all for 1pp4)
(see all for 1pp4)
Enzyme Mechanism
Introduction
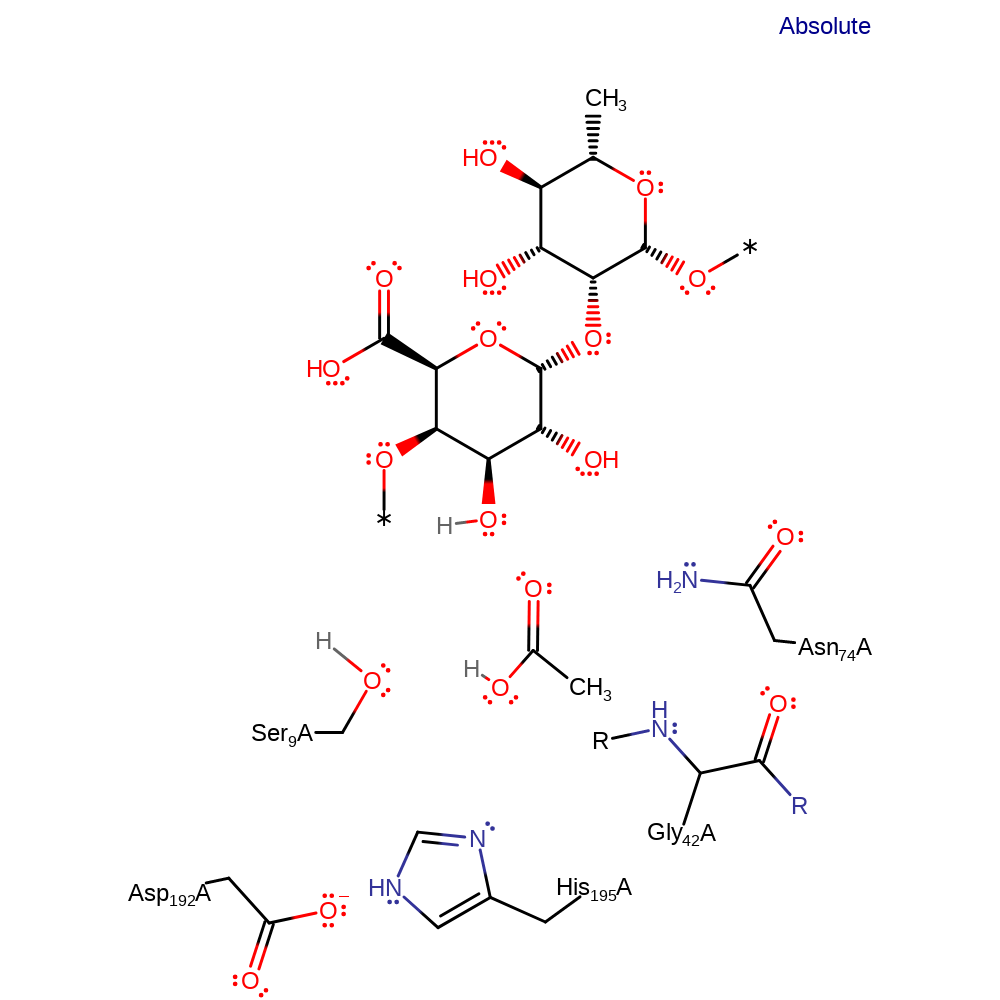
Serine-dependent hydrolases catalyse a two-step reaction involving acylation and deacylation, each proceeding via a tetrahedral intermediates. Asp-His both act to activate the nucleophilic serine by providing a proton sink. His195 deprotonates the Ser9 hydroxyl group, concomitantly with the nucleophilic attack of the serine residue on the carbonyl carbon of the substrate, forming a tetrahedral intermediate. This is stabilised by the formation of an oxyanion hole formed by hydrogen bonds to the main chain amides of Ser9 and Gly42, and the side chain amide of Asn74. The positively charged His195 donates a proton to the first leaving group, resulting in the collapse of the tetrahedral intermediate and the formation of the covalent acyl-enzyme intermediate. An incoming water molecule is deprotonated by His195. The activated water molecule nucleophilically attacks the central carbonyl carbon of the ester bond, hydrolysing the ester bond. This results in the formation and subsequent collapse of a tetrahedral intermediate, leading to the release of the product. The enzyme is regenerated by reprotonation of Ser9 by His195.
Catalytic Residues Roles
| UniProt | PDB* (1pp4) | ||
| Ser26 | Ser9A | Ser9 is the nucleophile that attacks the carbonyl carbon of the substrate. | covalently attached, nucleofuge, nucleophile, proton acceptor, proton donor, electrostatic stabiliser |
| Asn91, Ser26, Gly59 (main-N) | Asn74A, Ser9A, Gly42A (main-N) | The backbone amides of Gly42 and Ser9, and the sidechain amide of Asn74 form an oxyanion hole to stabilise the oxyanions formed during the reaction. | electrostatic stabiliser |
| Asp209 | Asp192A | Asp192 acts as a proton sink for Ser9 with His195. | increase basicity, electrostatic stabiliser |
| His212 | His195A | His195 deprotonates Ser9 and a water molecule to activate them as nucleophiles, and protonates the first leaving group and Ser9 when regenerating the enzyme. | proton acceptor, proton donor |
Chemical Components
overall reactant used, proton transfer, intermediate formation, bimolecular nucleophilic addition, overall product formed, unimolecular elimination by the conjugate base, intermediate terminated, native state of enzyme regeneratedReferences
- Mølgaard A et al. (2000), Structure, 8, 373-383. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. DOI:10.1016/s0969-2126(00)00118-0. PMID:10801485.
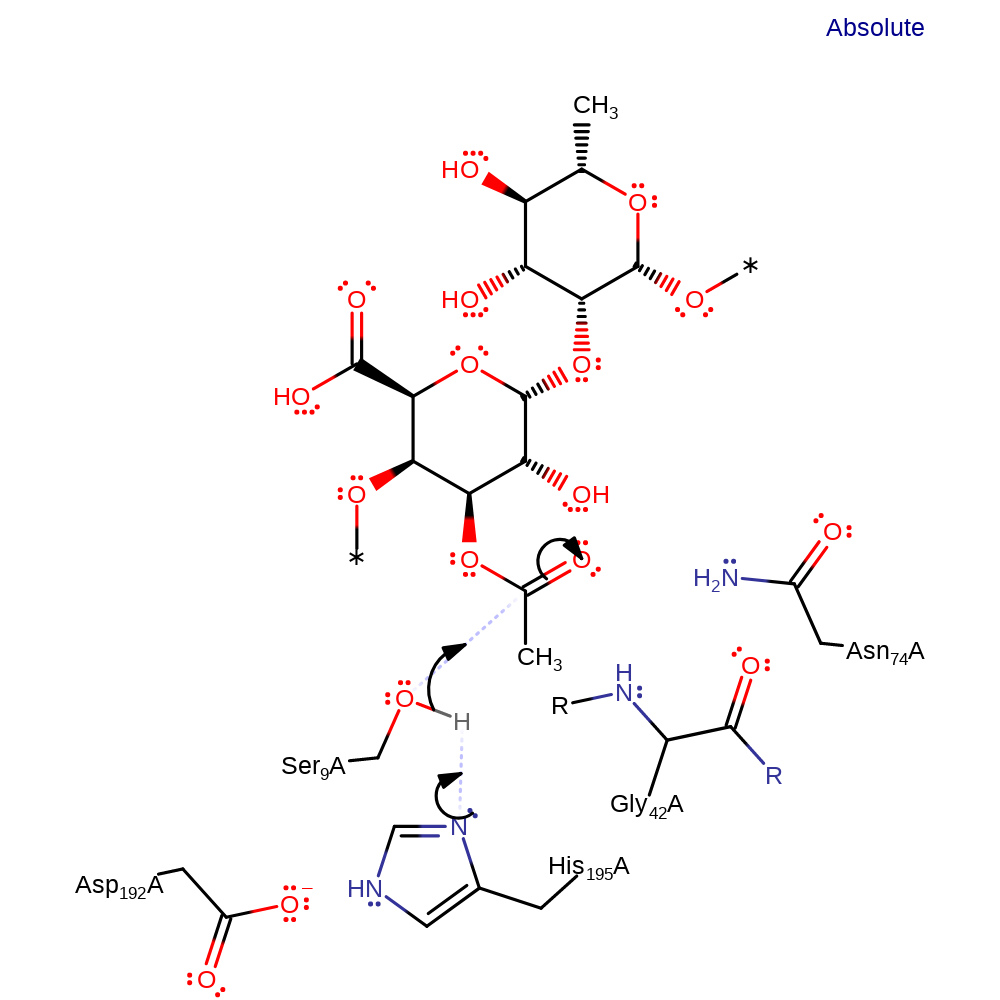
Step 1. His195 acts as a general base activating the Ser9 hydroxyl group for nucleophilic attack on the carbonyl carbon. The third component of this catalytic triad- Asp192 acts to increase the basicity of the histidine. The oxyanion intermediate formed is stabilized by the amide group of Gly42 and the side chain amide of Asn74.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly42A (main-N) | electrostatic stabiliser |
| Ser9A | electrostatic stabiliser, covalently attached |
| Asn74A | electrostatic stabiliser |
| Asp192A | increase basicity, electrostatic stabiliser |
| His195A | proton acceptor |
| Ser9A | proton donor, nucleophile |
Chemical Components
overall reactant used, proton transfer, intermediate formation, ingold: bimolecular nucleophilic addition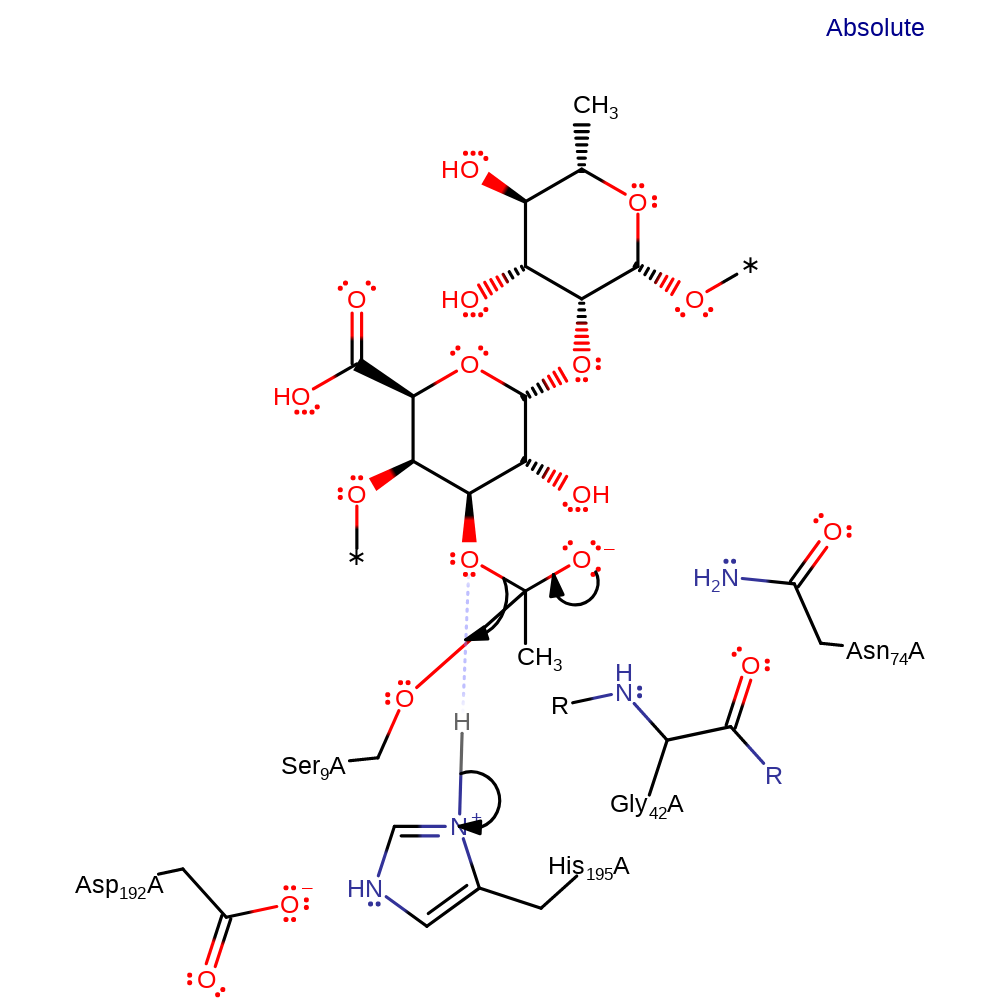
Step 2. The tetrahedral intermediate collapses and rhamnogalacturonan is eliminated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser9A | electrostatic stabiliser |
| Gly42A (main-N) | electrostatic stabiliser |
| Asn74A | electrostatic stabiliser |
| Asp192A | electrostatic stabiliser |
| Ser9A | covalently attached |
| His195A | proton donor |
Chemical Components
proton transfer, overall product formed, ingold: unimolecular elimination by the conjugate base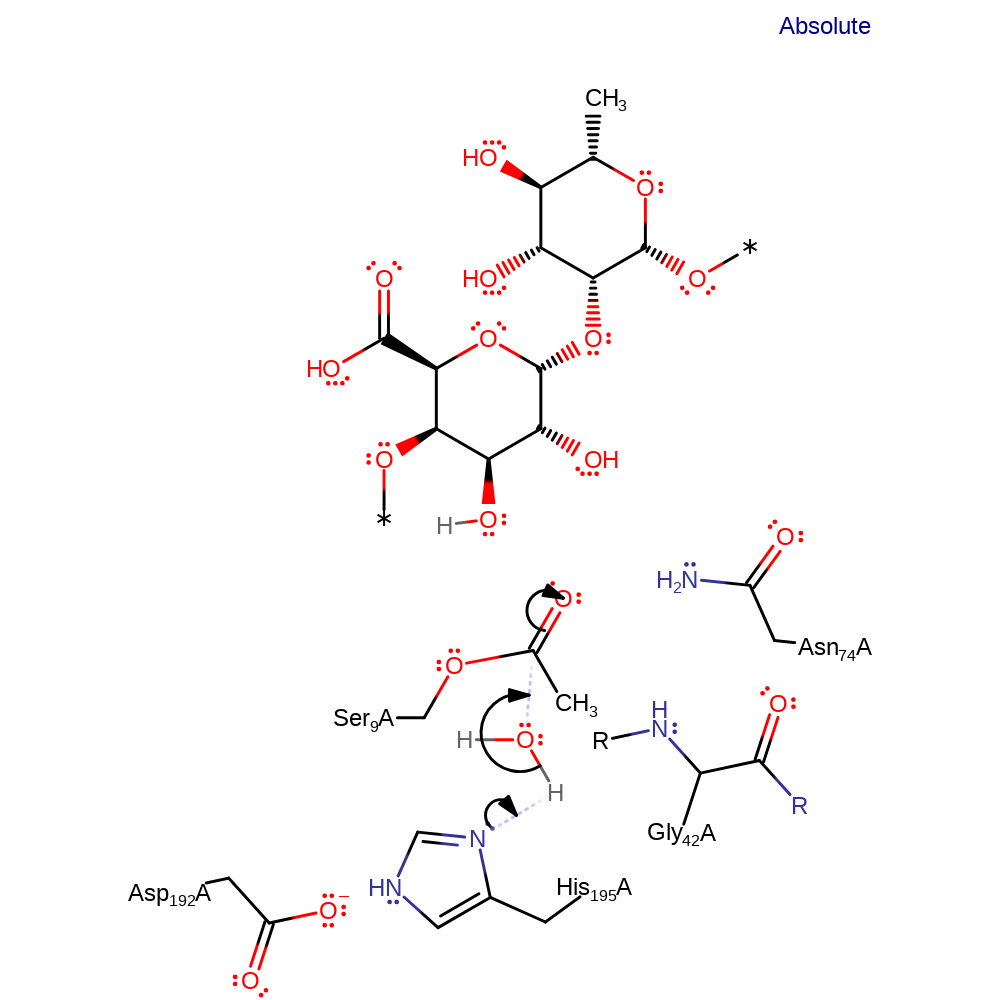
Step 3. His195 activates water for nucleophilic attack and another oxyanion intermediate is formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser9A | electrostatic stabiliser |
| Gly42A (main-N) | electrostatic stabiliser |
| Asn74A | electrostatic stabiliser |
| Asp192A | electrostatic stabiliser, increase basicity |
| Ser9A | covalently attached |
| His195A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic additionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser9A | electrostatic stabiliser |
| Gly42A (main-N) | electrostatic stabiliser |
| Asn74A | electrostatic stabiliser |
| Asp192A | electrostatic stabiliser |
| Ser9A | nucleofuge |
| His195A | proton donor |
| Ser9A | proton acceptor |





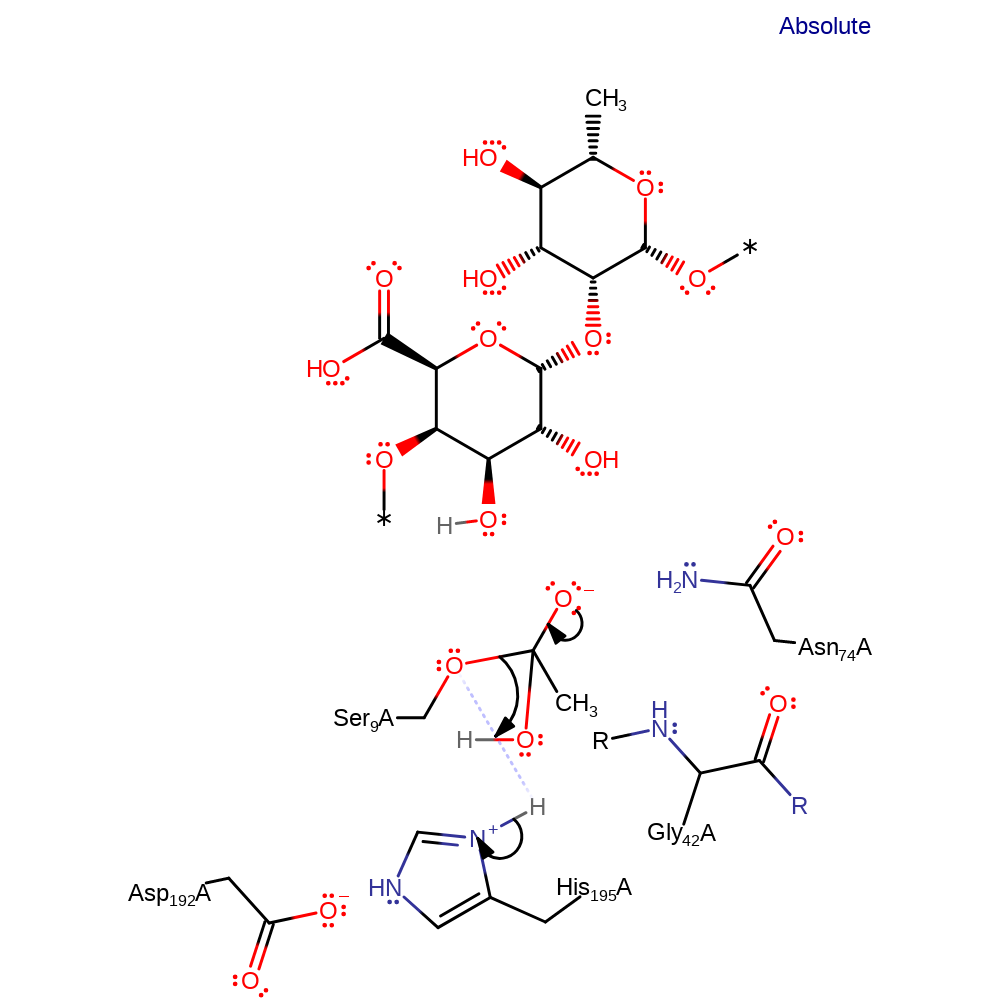
 Download:
Download: