Dihydroneopterin aldolase
7,8 dihydroneopterin from Staphylococcus aureus is able to catalyse the conversion of 7,8 dihydroneopterin to 6 hydroxymethyl 7,8 dihydroneopterin, an aldol cleavage reaction which also produces glycoaldehyde, a key step in the synthesis of folate, which can in turn be used for the synthesis of purines. The enzyme is an attractive target for antibiotics, as the it is found in MRSA, thus structural information can be used to design inhibitors of the enzyme.
Reference Protein and Structure
- Sequence
-
P56740
 (4.1.2.25, 5.1.99.8)
(4.1.2.25, 5.1.99.8)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Staphylococcus aureus (Bacteria)

- PDB
-
2dhn
- COMPLEX OF 7,8-DIHYDRONEOPTERIN ALDOLASE FROM STAPHYLOCOCCUS AUREUS WITH 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 2.2 A RESOLUTION
(2.2 Å)



- Catalytic CATH Domains
-
3.30.1130.10
 (see all for 2dhn)
(see all for 2dhn)
- Cofactors
- Water (1)
Enzyme Reaction (EC:4.1.2.25)
Enzyme Mechanism
Introduction
The reaction is an aldol cleavage reaction similar to those catalysed by aldolases. Deprotonation of the 7C OH by Lys 100, assisted by Glu 22, results in an unstable enamine intermediate which collapses to form the products glycoaldehyde and deprotonated 6 hydroxymethyl dho which picks up a proton from Lys 100 to regenerate the catalytically active form of the enzyme.
Catalytic Residues Roles
| UniProt | PDB* (2dhn) | ||
| Lys100 | Lys100A | Deprotonates the substrate thus forming the unstable enamine intermediate which collapses to release the products. Subsequently reprotonates the product to regenerate the active form of the enzyme. | proton acceptor, proton donor |
| Glu22 | Glu22A | Assists in the deprotonation of the substrate by maintaining Lys 100 in the correct protonation state through electrostatic contacts with the residue. | electrostatic stabiliser |
Chemical Components
proton transfer, overall product formed, overall reactant used, bimolecular elimination, assisted tautomerisation (not keto-enol), native state of enzyme regeneratedReferences
- Hennig M et al. (1998), Nat Struct Biol, 5, 357-362. Crystal structure and reaction mechanism of 7,8-dihydroneopterin aldolase from staphylococcus aureus. DOI:10.1038/nsb0598-357. PMID:9586996.
- Wang Y et al. (2006), Biochemistry, 45, 15232-15239. Mechanism of dihydroneopterin aldolase: functional roles of the conserved active site glutamate and lysine residues. DOI:10.1021/bi060949j. PMID:17176045.

Step 1. Lys100 deprotonates the C7 hydroxyl, this results in the C6-C7 bond being broken and an enamine intermediate being formed along with glycoaldehyde. Water acts as a general acid assisting in the enamine formation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu22A | electrostatic stabiliser |
| Lys100A | proton acceptor |
Chemical Components
proton transfer, overall product formed, overall reactant used, ingold: bimolecular elimination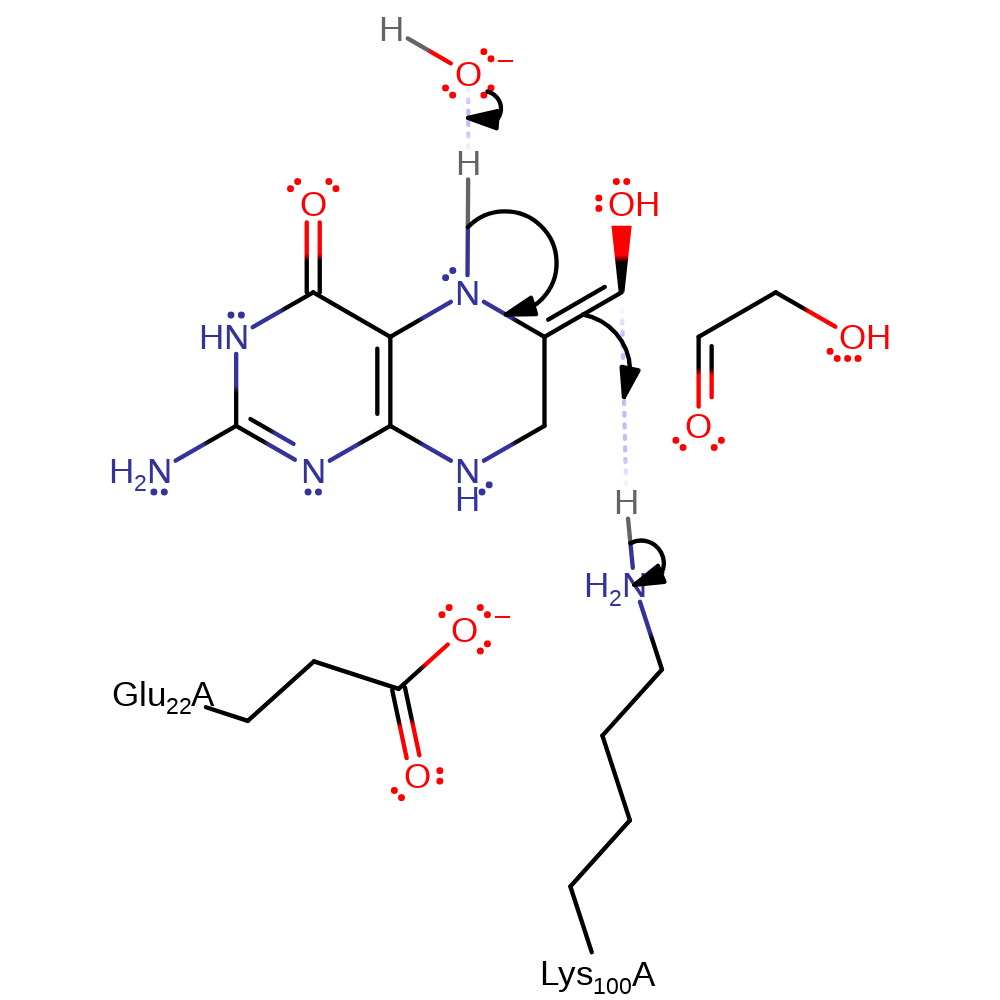
Step 2. The unstable enamine intermediate collapses upon protonation by Lys100 to form the product HP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys100A | proton donor |



 Download:
Download: