Bleomycin hydrolase
Bleomycin hydrolase is an aminopeptidase identified in eukaryotes ranging from yeast to mammals. It was originally discovered by its ability to hydrolyse the anti-cancer drug bleomycin, thus limiting the usefulness of this drug in the cell. The yeast enzyme (Gal6p) has been shown to negatively regulate the galactose metabolism system and to bind single-stranded DNA and RNA with high affinity; this binding coupled with the hydrolase activity has a role in the detoxification of bleomycin. Generation of the mature form of Gal6p involves the enzyme initially acting as a carboxypeptidase on its own C terminus to convert itself to an aminopeptidase.
Reference Protein and Structure
- Sequence
-
Q01532
 (3.4.22.40)
(3.4.22.40)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1gcb
- GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL)
(2.2 Å)



- Catalytic CATH Domains
-
3.90.70.10
 (see all for 1gcb)
(see all for 1gcb)
Enzyme Mechanism
Introduction
Bleomycin hydrolase is a cysteine protease. The resting state contains a thioloate-imidazolium pair (Cys 102 and His 398) that is stabilised by Asn 421. Cys 102 attacks the peptide carbonyl to form an acyl enzyme intermediate, with His 398 protonating the departing amine. Negative charge that accumulates on the substrate carbonyl oxygen during the reaction is stabilised by an 'oxyanion-hole' involving the side chain of Gln 96 and the backbone NH of Cys 102. The thioester intermediate is then hydrolysed by a water molecule that is deprotonated by His 398 acting as a general base.
Catalytic Residues Roles
| UniProt | PDB* (1gcb) | ||
| Cys102 | Cys73A | Attacks the peptide carbonyl to form an acyl-enzyme intermediate. Backbone NH forms part of the oxyanion hole and stabilises the negative charge that accumulates on the carbonyl oxygen in during the reaction. | nucleofuge, nucleophile, proton acceptor, proton donor, electrostatic stabiliser |
| Cys102 (main-N), Gln96 | Cys73A (main-N), Gln67A | Forms the oxyanion hole which functions to stabilise the negative charge that accumulates on the carbonyl oxygen of the substrate during the reaction. | electrostatic stabiliser |
| His398 | His369A | Deprotonates Cys 102 to give the thiolate which acts as a nucleophile to attack the peptide bond. Protonates the departing amine group of the substrate. Later acts as a general base to deprotonate a water molecule that attacks the thioester intermediate. | proton acceptor, proton donor |
| Asn421 | Asn392A | Stabilises the thiolate-imidazolium pair by interacting with His 398. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Joshua-Tor L et al. (1995), Science, 269, 945-950. Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6. DOI:10.1126/science.7638617. PMID:7638617.
- O'Farrell PA et al. (2007), Biochem J, 401, 421-428. Mutagenesis and crystallographic studies of the catalytic residues of the papain family protease bleomycin hydrolase: new insights into active-site structure. DOI:10.1042/BJ20060641. PMID:17007609.
- Lefterov IM et al. (2001), Biochem Biophys Res Commun, 283, 994-999. Cysteine 73 in bleomycin hydrolase is critical for amyloid precursor protein processing. DOI:10.1006/bbrc.2001.4860. PMID:11350084.
- O’Farrell PA et al. (1999), Structure, 7, 619-627. Crystal structure of human bleomycin hydrolase, a self-compartmentalizing cysteine protease. DOI:10.1016/S0969-2126(99)80083-5.
- Zheng W et al. (1998), Cell, 93, 103-109. The Unusual Active Site of Gal6/Bleomycin Hydrolase Can Act as a Carboxypeptidase, Aminopeptidase, and Peptide Ligase. DOI:10.1016/s0092-8674(00)81150-2. PMID:9546396.

Step 1. His398 deprotonates Cys102 which activates Cys102 to attack the carbon of the carbonyl in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn392A | electrostatic stabiliser |
| Cys73A | electrostatic stabiliser |
| Gln67A | electrostatic stabiliser |
| Cys73A | proton donor, nucleophile |
| His369A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 2. The oxyanion initiates an elimination which results in the cleavage of the peptide bond. The amino group of the N-terminal product accepts a proton from His398.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln67A | electrostatic stabiliser |
| Cys73A (main-N) | electrostatic stabiliser |
| Asn392A | electrostatic stabiliser |
| His369A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, overall product formed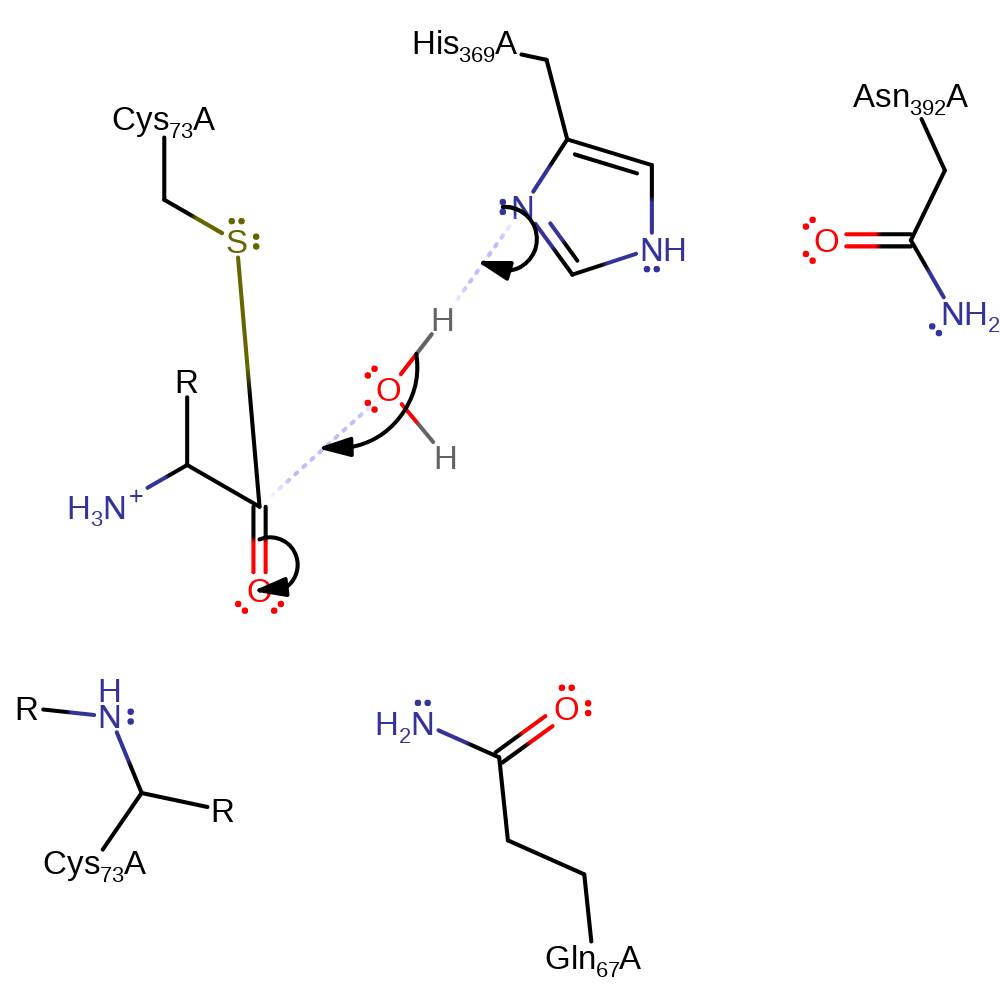
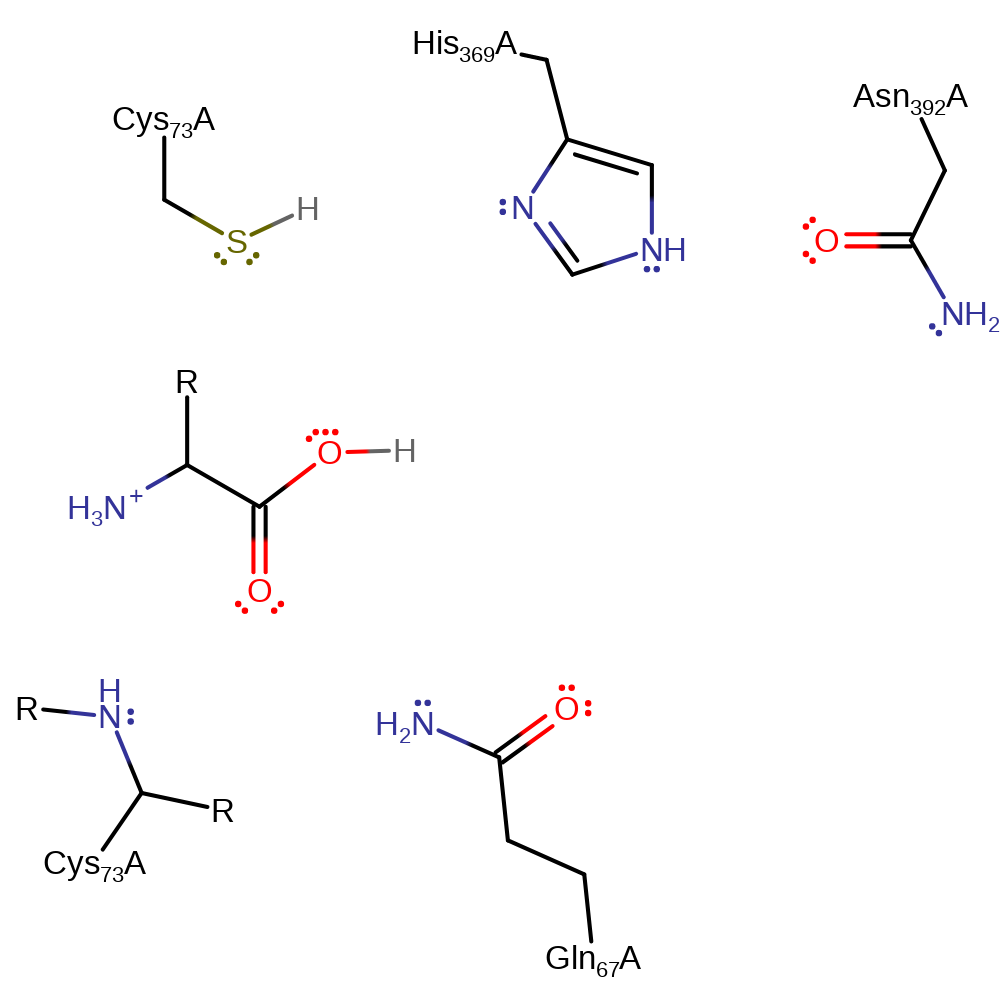
Step 3. His398 deprotonates a water which activates it so it can nucleophilically attack the carbon of the thioester bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln67A | electrostatic stabiliser |
| Cys73A (main-N) | electrostatic stabiliser |
| Asn392A | electrostatic stabiliser |
| His369A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used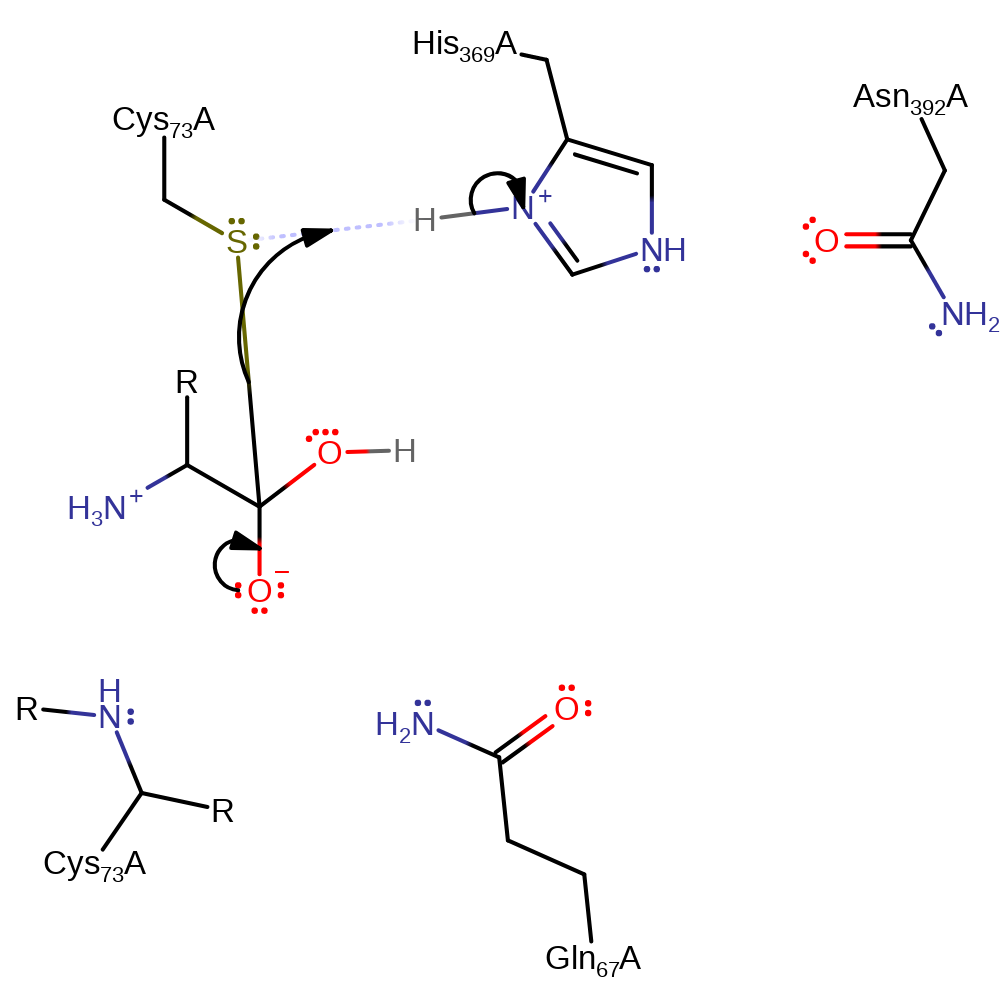
Step 4. The oxyanion initiates another elimination which results in the cleavage of the thioester bond. The released Cys102 now accepts a proton from His398 which returns the enzyme back to its native state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln67A | electrostatic stabiliser |
| Cys73A (main-N) | electrostatic stabiliser |
| Asn392A | electrostatic stabiliser |
| Cys73A | proton acceptor, nucleofuge |
| His369A | proton donor |



 Download:
Download: