Anhydrosialidase
Intramolecular trans-sialidase (IT-sialidase) specifically cleaves the terminal N-acetyl-neuraminic acid-alpha-2-3-Galactose linkage in sialoglyco-conjugates. The released product is 2,7-anhydro-N-acetyl-neuraminic acid, indicating that the enzyme carries out an intramolecular reaction mechanism. This is in contrast to the hydrolytic sialidases whose product is free N-acetyl-neuraminic acid.
Reference Protein and Structure
- Sequence
-
Q27701
 (4.2.2.15)
(4.2.2.15)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Macrobdella decora (North American leech)

- PDB
-
1sll
- SIALIDASE L FROM LEECH MACROBDELLA DECORA
(2.0 Å)



- Catalytic CATH Domains
-
2.120.10.10
 (see all for 1sll)
(see all for 1sll)
Enzyme Reaction (EC:4.2.2.15)
Enzyme Mechanism
Introduction
In the first step, O2 of the substrate acquires a proton and leaves. Asp 318 either provides this proton directly or stabilises a proton from the solvent in the vicinity. The resulting positively charged oxocarbonium intermediate is stabilised by the negative side chains of Asp 318 and Glu 595, and also by a lone pair from the phenolic OH of Tyr 713. In the second step, the C7 hydroxyl attacks the oxocarbonium ion at C2. Asp 318 acts as a general base to deprotonate the attacking hydroxyl group.
Catalytic Residues Roles
| UniProt | PDB* (1sll) | ||
| Asp318 | Asp318(238)A | May protonate departing O2 of substrate, or at least stabilise a proton in the vicinity. Provides electrostatic stabilisation for the positively charged oxocarbonium ion. Accepts proton from the O7 hydroxyl group that attacks the oxocarbonium ion. | proton acceptor, electrostatic stabiliser, proton donor |
| Glu595 | Glu595(515)A | Provides electrostatic stabilisation for the positively charged oxocarbonium ion. Also interacts with Tyr 713 to direct its O lone pair towards the substrate C2. | electrostatic stabiliser |
| Tyr713 | Tyr713(633)A | Stabilises the oxocarbonium ion by directing a lone pair on the phenolic oxygen towards C2 of the substrate. | electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, overall product formed, heterolysis, intramolecular nucleophilic addition, native state of enzyme regeneratedReferences
- Luo Y et al. (1999), J Mol Biol, 285, 323-332. The 1.8 å structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism. DOI:10.1006/jmbi.1998.2345. PMID:9878409.
- Cremona ML et al. (1995), Gene, 160, 123-128. A single tyrosine differentiates active and inactive Trypanosoma cruzi trans-sialidases. DOI:10.1016/0378-1119(95)00175-6. PMID:7628705.
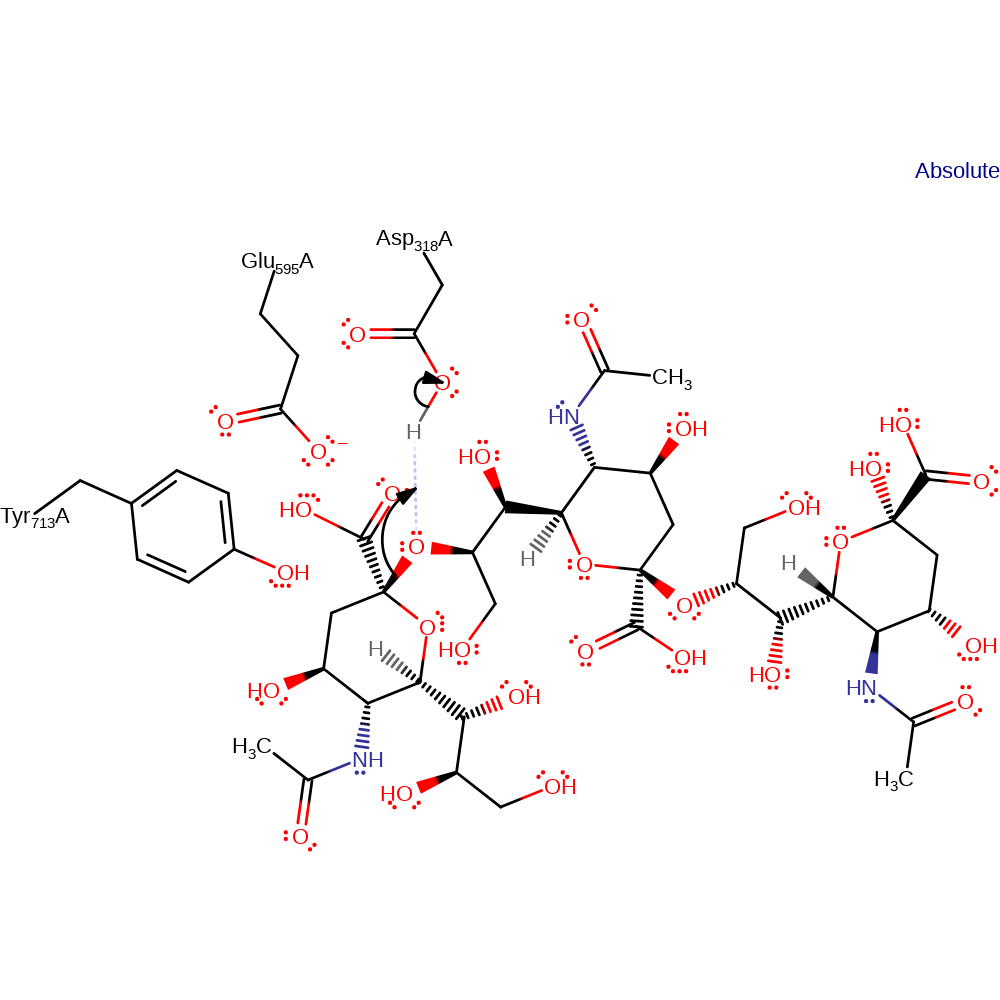
Step 1. Asp 318 protonates O2 of the substrate this causes the C2-O2 bond to be broken. The resulting oxocarbonium intermediate is stabilized by Asp 318, Glu595 and Tyr713.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu595(515)A | electrostatic stabiliser |
| Tyr713(633)A | electrostatic stabiliser |
| Asp318(238)A | electrostatic stabiliser, proton donor |
Chemical Components
proton transfer, overall reactant used, overall product formed, heterolysis
Step 2. The C7 hydroxyl attacks the oxocarbonium ion at C2, Asp 318 acts as a general base deprotonating the hydroxyl. Leading to the second product being formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp318(238)A | electrostatic stabiliser |
| Glu595(515)A | electrostatic stabiliser |
| Tyr713(633)A | electrostatic stabiliser |
| Asp318(238)A | proton acceptor |



 Download:
Download: