[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase
Ribulose-1,5-bisphosphate carboxylase-oxygenase large subunit N-methyltransferase enzymes (LSMTs) uses S-adenosyl methionine to methylate Lys 14 of the Rubisco large subunit. Lys 14 is situated in the flexible N-terminal tail of the large subunit, and the precise role of its methylation is not clear. It does not affect the activity of Rubisco but may be involved in targeting other proteins to interact with the tail.
Reference Protein and Structure
- Sequence
-
Q43088
 (2.1.1.127, 2.1.1.259)
(2.1.1.127, 2.1.1.259)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pisum sativum (pea)

- PDB
-
1mlv
- Structure and Catalytic Mechanism of a SET Domain Protein Methyltransferase
(2.6 Å)



- Catalytic CATH Domains
-
3.90.1410.10
 (see all for 1mlv)
(see all for 1mlv)
Enzyme Reaction (EC:2.1.1.127)
Enzyme Mechanism
Introduction
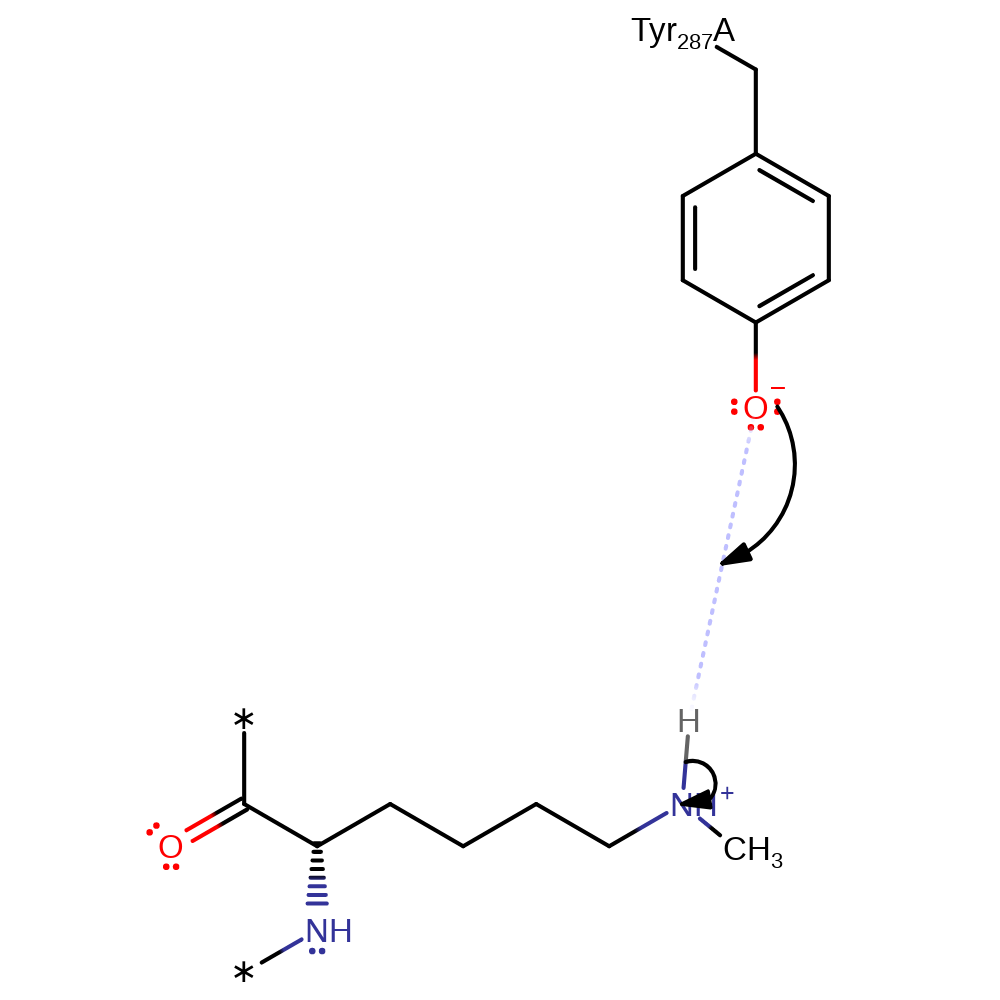
The ionised form of Tyr 287 deprotonates the -NH3+ group of Lys 14 (on the large subunit of Rubisco). The resulting Lys-NH2 group can then attack the methyl group of the AdoMet sulphonium cation. It is proposed that the pKa of Tyr 287 is depressed (so allowing it to be present in the ionised form which can act as a general base) by the proximity of two positively charged groups: the AdoMet sulfonium cation and the epsilon-ammonium group of the substrate lysine.
Catalytic Residues Roles
| UniProt | PDB* (1mlv) | ||
| Tyr287 | Tyr287(243)A | Removes proton from Lys14-NH3+ to generate the nucleophilic species Lys14-NH2. | activator, proton acceptor, proton donor |
Chemical Components
proton transfer, overall reactant used, bimolecular nucleophilic substitution, intramolecular nucleophilic substitution, overall product formed, inferred reaction step, native state of enzyme regeneratedReferences
- Trievel RC et al. (2002), Cell, 111, 91-103. Structure and catalytic mechanism of a SET domain protein methyltransferase. DOI:10.2210/pdb1mlv/pdb. PMID:12372303.
- Zhang X et al. (2007), Biochemistry, 46, 5505-5514. Catalytic mechanism and product specificity of rubisco large subunit methyltransferase: QM/MM and MD investigations. DOI:10.1021/bi700119p. PMID:17429949.

Step 1. The unprotonated form of Tyr287 deprotonates the ammonium group of Lys14.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr287(243)A | activator, proton acceptor |
Chemical Components
proton transfer, overall reactant used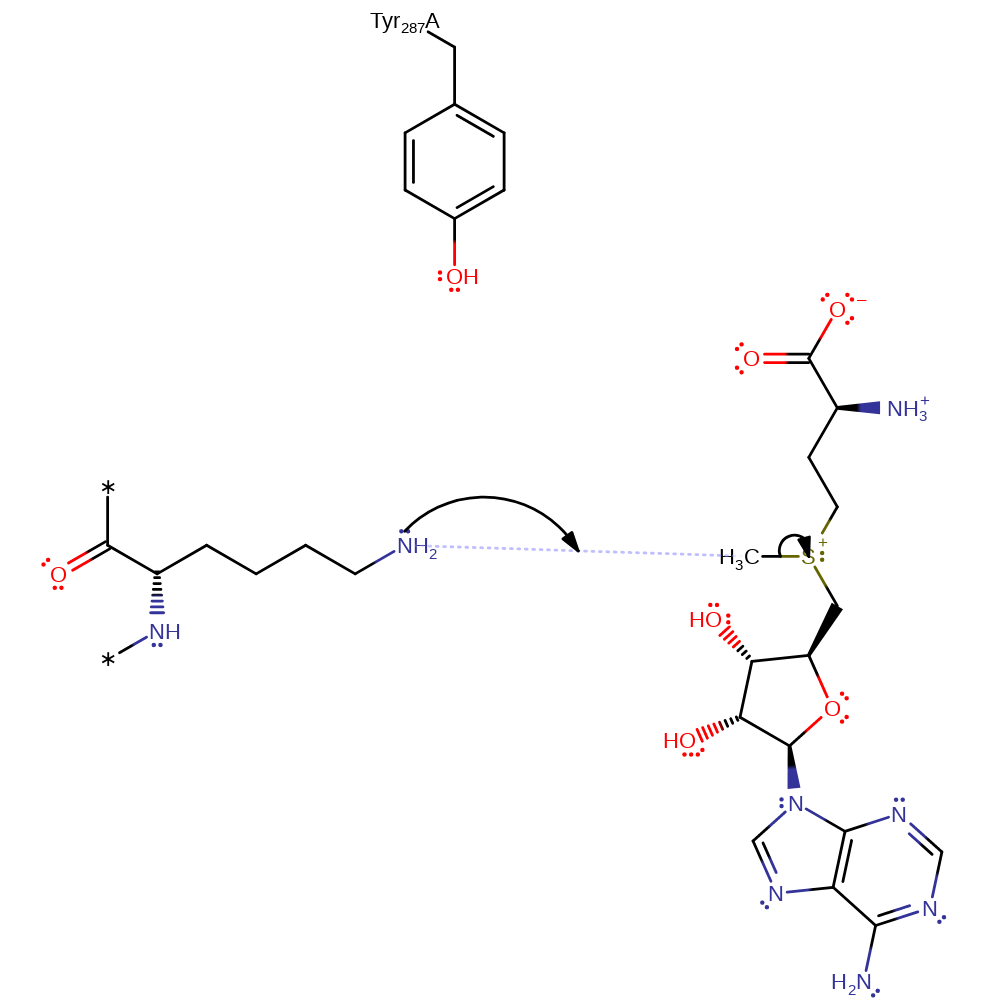
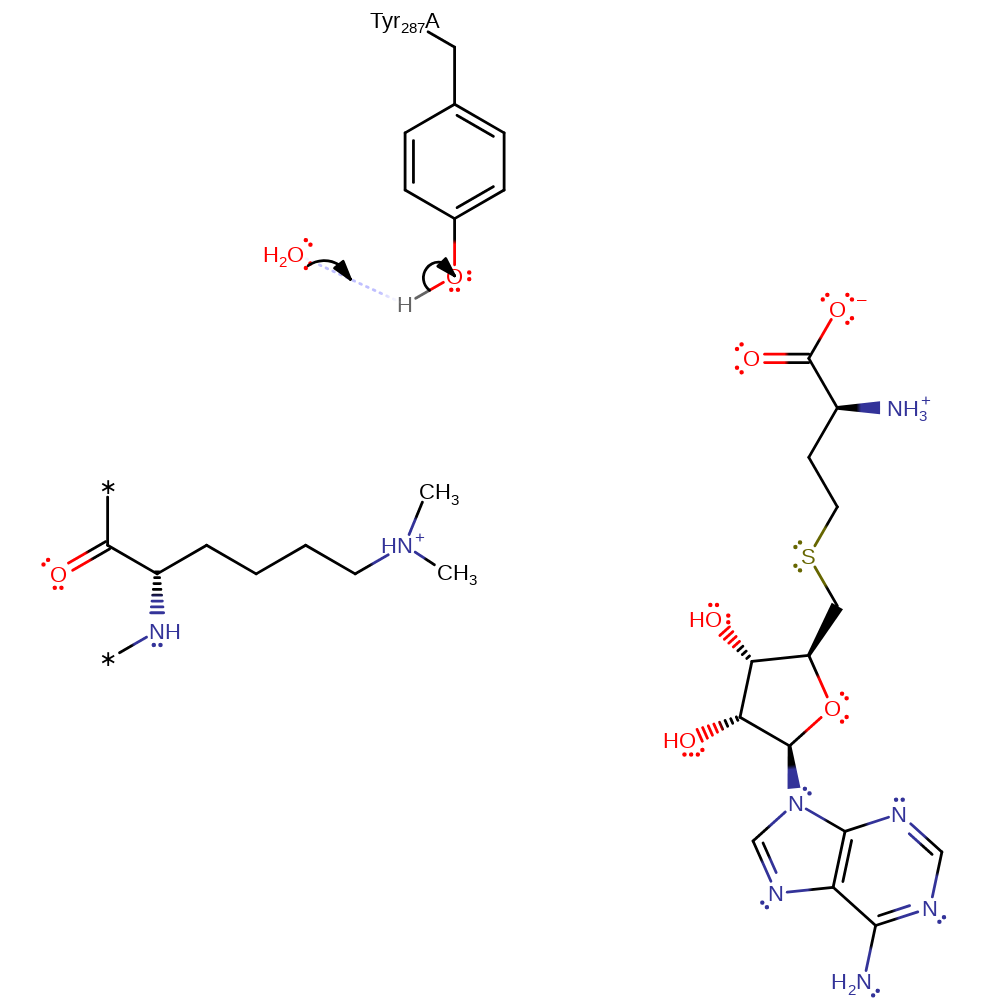
Step 2. In an SN2 reaction the primary amine group of Lys14 attacks the methyl group of SAM.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic substitutionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr287(243)A | proton donor |
Chemical Components
proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr287(243)A | activator, proton acceptor |
Chemical Components
proton transfer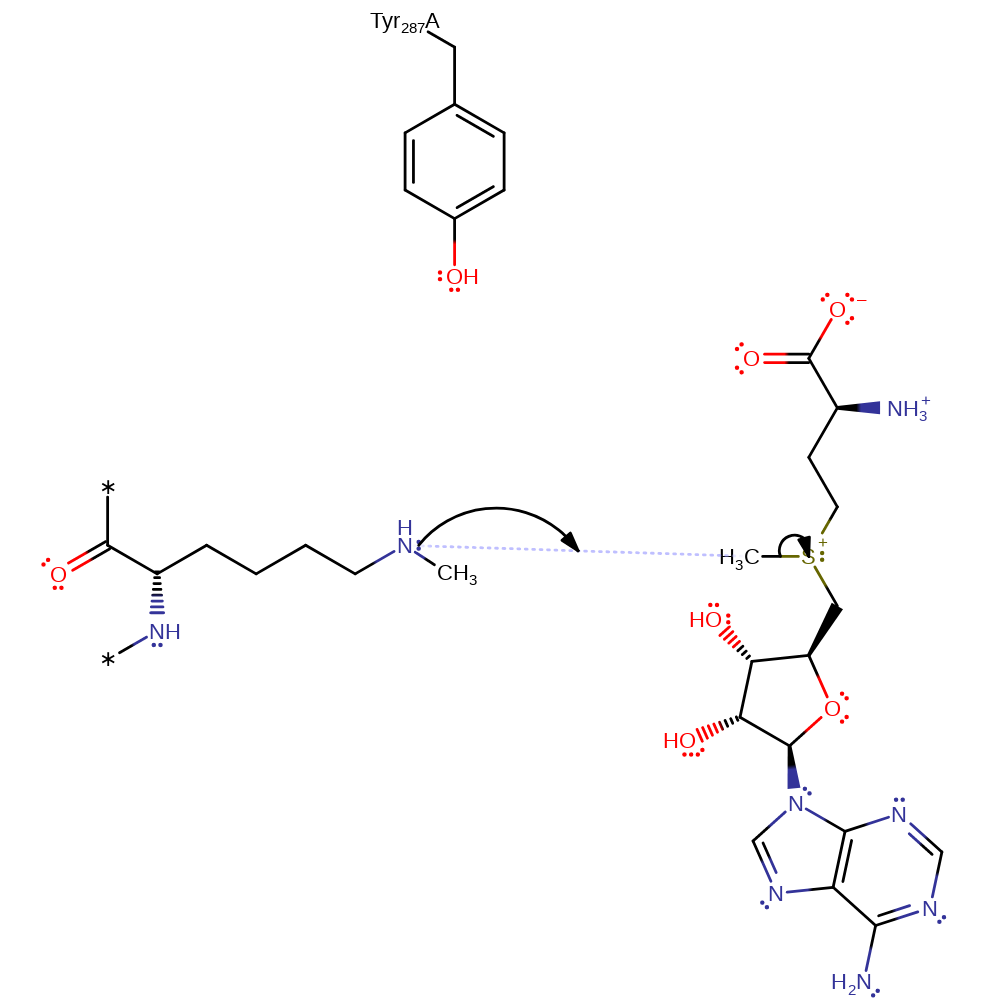
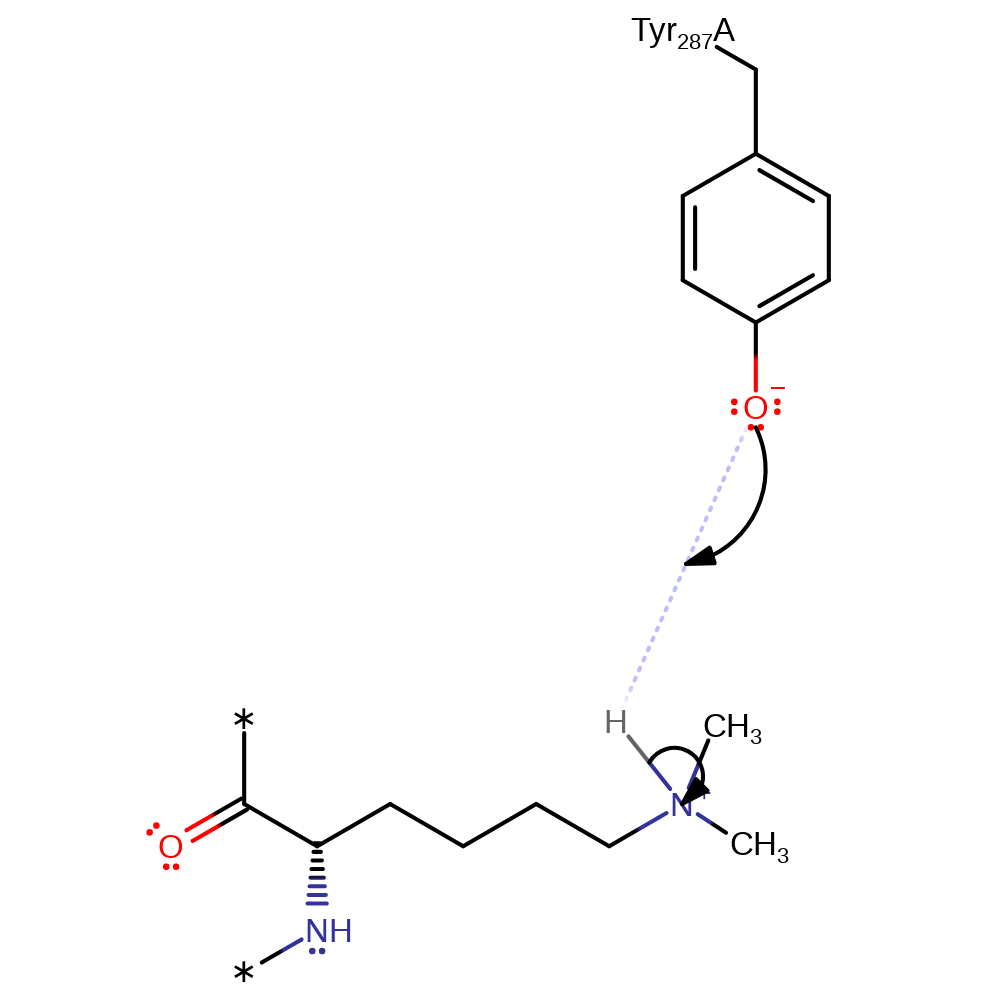
Step 5. In an SN2 reaction Lys14 attacks the methyl group of SAM to form a tertiary amine and SAH.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic substitutionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr287(243)A | proton donor |
Chemical Components
proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr287(243)A | activator, proton acceptor |
Chemical Components
proton transfer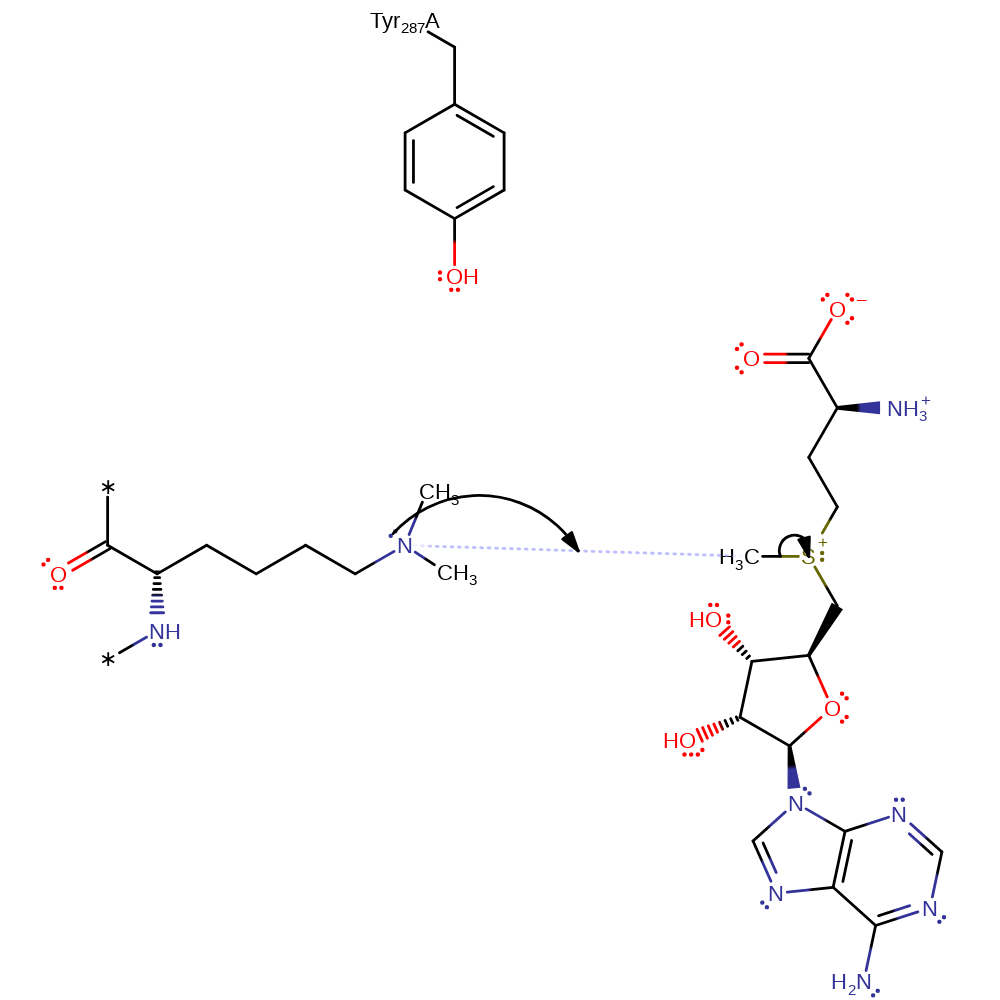
Step 8. In an SN2 reaction the tertiary amine of Lys14 attacks SAM, forming the quaternary amine product and SAH.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: intramolecular nucleophilic substitution, overall product formed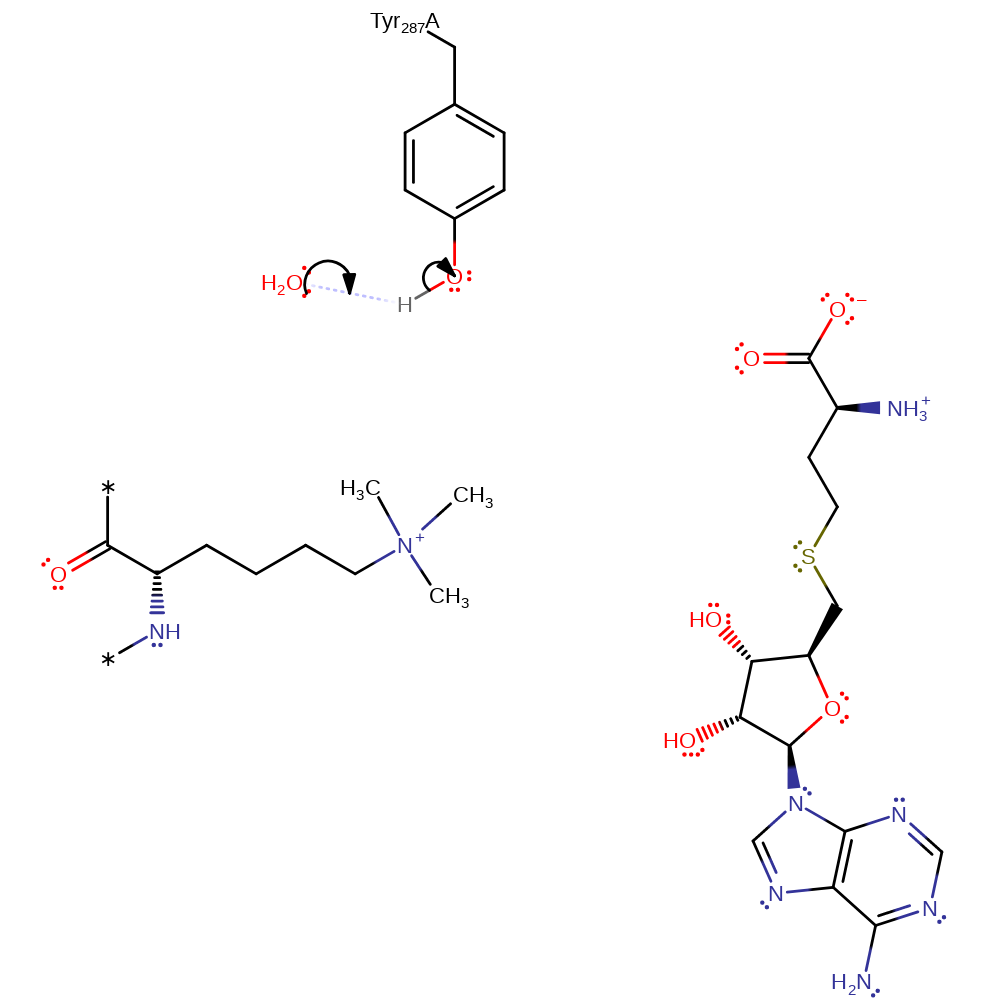
Step 9. In an inferred reaction step Tyr287 is deprotonated to regenerate the native state of the enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr287(243)A | proton donor |






 Download:
Download: