Thymidine kinase
Thymidine kinases (TK) are key enzymes in the pyrimidine salvage pathway catalyzing the phosphate transfer from ATP to thymidine (dT) in the presence of Mg2+ and thus, yielding thymidine monophosphate (dTMP) and ADP. Herpesviridae, such as Herpes simplex virus type 1, encode for their own, multifunctional TK. Unlike the very specific human cytosolic TK (TK1), it is able to phosphorylate pyrimidine as well as purine analogs and demands less stereochemical restrictions concerning the sugar moiety also accepting acyclic side chains as phosphate acceptors (e.g., aciclovir). Therefore, the difference in substrate specificity of human TK 1 and TKHSV1 is a crucial point in establishing a molecular basis for selective antiviral therapy, featuring TKHSV1 as the center of activation of antiviral drugs such as aciclovir (ACV), penciclovir, and ganciclovir (GCV). First being activated by phosphorylation by viral encoded TK, these nucleoside analogs in their triphosphate form block the viral replication by subsequently terminating DNA elongation at the viral DNA polymerase. In combination with GCV TKHSV1 is an established tool used as a prodrug-activating enzyme, so-called suicide enzyme, in gene therapy of cancer, AIDS, and in controlling graft-versus-host disease by allogenic bone marrow transplant (allo BMT).
Reference Protein and Structure
- Sequence
-
P03176
 (2.7.1.21)
(2.7.1.21)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Herpes simplex virus (type 1 / strain 17) (Virus)

- PDB
-
1kim
- CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH DEOXYTHYMIDINE
(2.14 Å)



- Catalytic CATH Domains
-
3.40.50.300
 (see all for 1kim)
(see all for 1kim)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.7.1.21)
Enzyme Mechanism
Introduction
The catalytic mechanism proceeds as follows: Glu83 acts as a general base to activate the acceptor group, 5'-hydroxyl group of thymidine. The activated acceptor group makes a nucleophilic attack on the transferred group, gamma-phosphate of ATP. During the transition state, the transferred group and leaving group (beta- and alpha-phosphate groups of ATP) must be stabilised by stabiliser residues, Arg/Lys cluster, along with the magnesium ion as cofactor. It is proposed Asp162 can bind to ATP or ADP through the magnesium ion, although the exact position and interactions of magnesium are unclear from published protein structures.
Catalytic Residues Roles
| UniProt | PDB* (1kim) | ||
| Asp162 | Asp162(152)A | Proposed to coordinate to a magnesium ion. | metal ligand |
| Glu225 | Glu225(215)A | Important dipole interaction with sugar moiety | electrostatic stabiliser, polar interaction |
| Arg220, Arg222 | Arg220(210)A, Arg222(212)A | Forms an anion hole to make the phosphate atom more electrophilic. | electrostatic stabiliser, polar interaction |
| Arg163 | Arg163(153)A | Stabilise transition group to aid transfer. Also could position the thymidine for phosphorylation. | electrostatic stabiliser, polar interaction |
| Lys62 | Lys62(52)A | Lys62 was suggested to aid the gamma-phosphoryl transfer during catalysis via stabilisation of the transition state. | electrostatic stabiliser, polar interaction |
| Glu83 | Glu83(73)A | Acts as a general base to deprotonate the O-5' atom to increase its nucleophilicity and activate it for attack. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, inferred reaction step, overall product formed, native state of enzyme regeneratedReferences
- Sulpizi M et al. (2001), J Biol Chem, 276, 21692-21697. The Rational of Catalytic Activity of Herpes Simplex Virus Thymidine Kinase: A COMBINED BIOCHEMICAL AND QUANTUM CHEMICAL STUDY. DOI:10.1074/jbc.m010223200. PMID:11262392.
- Gardberg A et al. (2003), Structure, 11, 1265-1277. Structural Basis for the Dual Thymidine and Thymidylate Kinase Activity of Herpes Thymidine Kinases. DOI:10.1016/j.str.2003.09.003.
- Champness JN et al. (1998), Proteins, 32, 350-361. Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands. DOI:10.1002/(sici)1097-0134(19980815)32:3<350::aid-prot10>3.0.co;2-8. PMID:9715911.
- Wild K et al. (1997), Protein Sci, 6, 2097-2106. The structures of thymidine kinase from Herpes simplex virus type 1 in complex with substrates and a substrate analogue. DOI:10.1002/pro.5560061005. PMID:9336833.
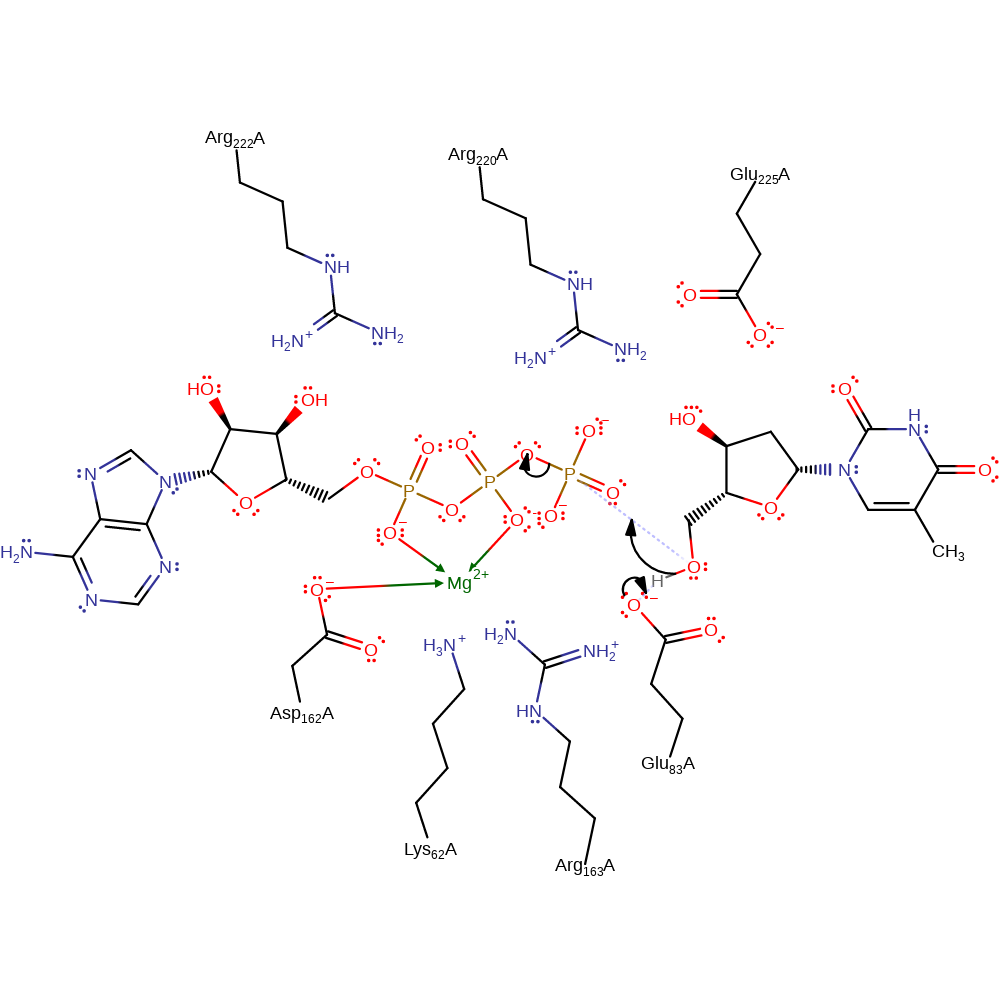
Step 1. Glu83 acts as a base catalyst and deprotonates thymidines 5' OH group. This increases the OH group's nucleophilicity to attack the gamma phosphate on ATP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg222(212)A | electrostatic stabiliser |
| Arg220(210)A | electrostatic stabiliser |
| Arg163(153)A | electrostatic stabiliser |
| Lys62(52)A | electrostatic stabiliser |
| Glu225(215)A | electrostatic stabiliser |
| Asp162(152)A | metal ligand |
| Lys62(52)A | polar interaction |
| Arg163(153)A | polar interaction |
| Arg220(210)A | polar interaction |
| Arg222(212)A | polar interaction |
| Glu225(215)A | polar interaction |
| Glu83(73)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used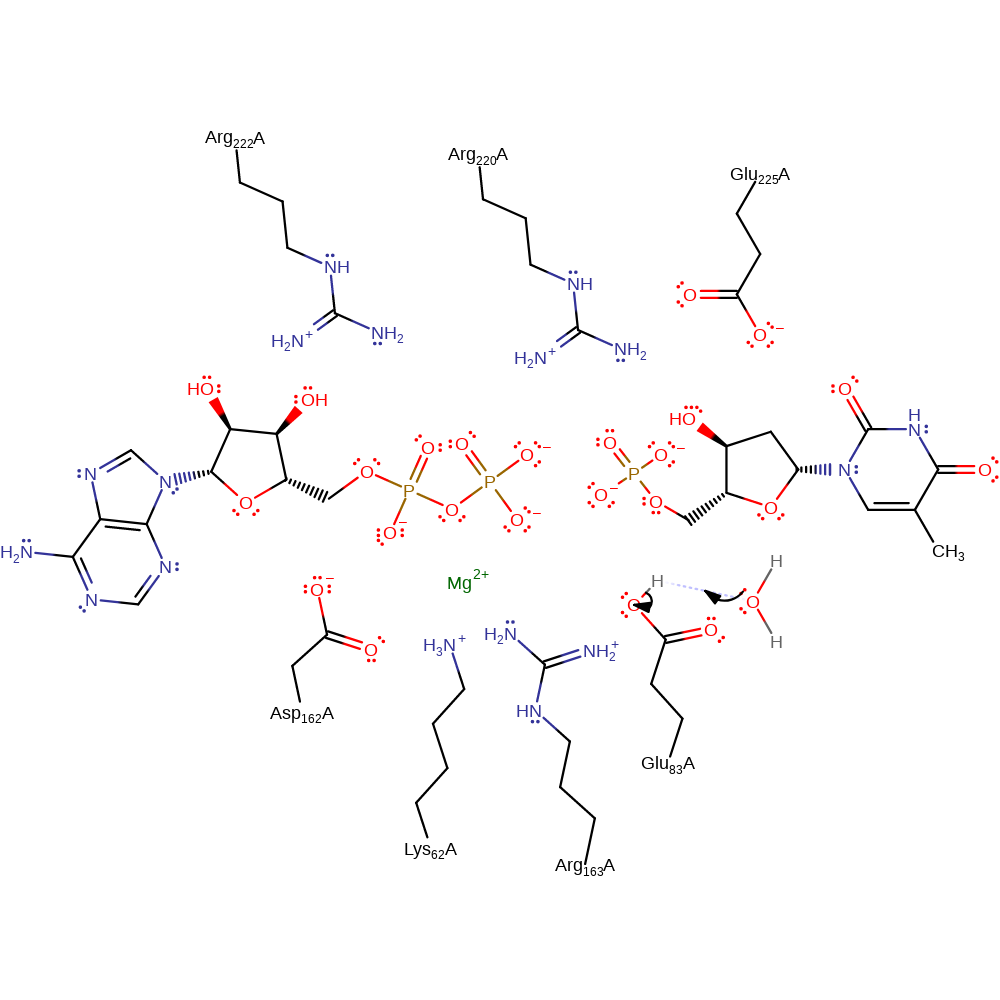
Step 2. Inferred deprotonation of Glu83 ready for the next round of catalysis.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu83(73)A | proton donor |





 Download:
Download: