 |
PDBsum entry 1kim

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of thymidine kinase from herpes simplex virus type i complexed with deoxythymidine
|
|
Structure:
|
 |
Thymidine kinase. Chain: a, b. Synonym: tk. Engineered: yes
|
|
Source:
|
 |
Herpes simplex virus (type 1 / strain 17). Organism_taxid: 10299. Strain: 17. Gene: tk. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.14Å
|
R-factor:
|
0.209
|
R-free:
|
0.279
|
|
|
Authors:
|
 |
J.N.Champness,M.S.Bennett,F.Wien,D.G.Brown,R.Visse,G.Sandhu,A.Davies, P.J.Rizkallah,C.Melitz,W.C.Summers,M.R.Sanderson
|
|
Key ref:
|
 |
J.N.Champness
et al.
(1998).
Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins,
32,
350-361.
PubMed id:
|
 |
|
Date:
|
 |
|
12-Nov-97
|
Release date:
|
20-May-98
|
|
|
Supersedes:
|

|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0DTH5
(KITH_HHV11) -
Thymidine kinase from Human herpesvirus 1 (strain 17)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
376 a.a.
304 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.21
- thymidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
thymidine + ATP = dTMP + ADP + H+
|
 |
 |
 |
 |
 |
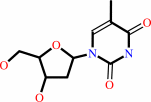 thymidine
thymidine
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 dTMP
dTMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Proteins
32:350-361
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
|
|
J.N.Champness,
M.S.Bennett,
F.Wien,
R.Visse,
W.C.Summers,
P.Herdewijn,
E.de Clerq,
T.Ostrowski,
R.L.Jarvest,
M.R.Sanderson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Antiherpes therapies are principally targeted at viral thymidine kinases and
utilize nucleoside analogs, the triphosphates of which are inhibitors of viral
DNA polymerase or result in toxic effects when incorporated into DNA. The most
frequently used drug, aciclovir (Zovirax), is a relatively poor substrate for
thymidine kinase and high-resolution structural information on drugs and other
molecules binding to the target is therefore important for the design of novel
and more potent chemotherapy, both in antiherpes treatment and in gene therapy
systems where thymidine kinase is expressed. Here, we report for the first time
the binary complexes of HSV-1 thymidine kinase (TK) with the drug molecules
aciclovir and penciclovir, determined by X-ray crystallography at 2.37 A
resolution. Moreover, from new data at 2.14 A resolution, the refined structure
of the complex of TK with its substrate deoxythymidine (R = 0.209 for 96% of all
data) now reveals much detail concerning substrate and solvent interactions with
the enzyme. Structures of the complexes of TK with four halogen-containing
substrate analogs have also been solved, to resolutions better than 2.4 A. The
various TK inhibitors broadly fall into three groups which together probe the
space of the enzyme active site in a manner that no one molecule does alone, so
giving a composite picture of active site interactions that can be exploited in
the design of novel compounds.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.C.Hsu,
Y.F.Chen,
S.R.Lin,
and
J.M.Yang
(2011).
iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis.
|
| |
BMC Bioinformatics,
12,
S33.
|
 |
|
|
|
|
 |
D.L.Clinciu,
Y.F.Chen,
C.N.Ko,
C.C.Lo,
and
J.M.Yang
(2010).
TSCC: Two-Stage Combinatorial Clustering for virtual screening using protein-ligand interactions and physicochemical features.
|
| |
BMC Genomics,
11,
S26.
|
 |
|
|
|
|
 |
Y.F.Chen,
K.C.Hsu,
S.R.Lin,
W.C.Wang,
Y.C.Huang,
and
J.M.Yang
(2010).
SiMMap: a web server for inferring site-moiety map to recognize interaction preferences between protein pockets and compound moieties.
|
| |
Nucleic Acids Res,
38,
W424-W430.
|
 |
|
|
|
|
 |
Y.Mehellou,
J.Balzarini,
and
C.McGuigan
(2009).
An investigation into the anti-HIV activity of 2',3'-didehydro-2',3'-dideoxyuridine (d4U) and 2',3'-dideoxyuridine (ddU) phosphoramidate 'ProTide' derivatives.
|
| |
Org Biomol Chem,
7,
2548-2553.
|
 |
|
|
|
|
 |
I.T.Hussein,
R.N.Miguel,
L.S.Tiley,
and
H.J.Field
(2008).
Substrate specificity and molecular modelling of the feline herpesvirus-1 thymidine kinase.
|
| |
Arch Virol,
153,
495-505.
|
 |
|
|
|
|
 |
N.E.Mikkelsen,
B.Munch-Petersen,
and
H.Eklund
(2008).
Structural studies of nucleoside analog and feedback inhibitor binding to Drosophila melanogaster multisubstrate deoxyribonucleoside kinase.
|
| |
FEBS J,
275,
2151-2160.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Candice L,
S.Django,
and
B.Margaret E
(2008).
The role of herpes simplex virus-1 thymidine kinase alanine 168 in substrate specificity.
|
| |
Open Biochem J,
2,
60-66.
|
 |
|
|
|
|
 |
V.Ramensky,
A.Sobol,
N.Zaitseva,
A.Rubinov,
and
V.Zosimov
(2007).
A novel approach to local similarity of protein binding sites substantially improves computational drug design results.
|
| |
Proteins,
69,
349-357.
|
 |
|
|
|
|
 |
J.Balzarini,
S.Liekens,
N.Solaroli,
K.El Omari,
D.K.Stammers,
and
A.Karlsson
(2006).
Engineering of a single conserved amino acid residue of herpes simplex virus type 1 thymidine kinase allows a predominant shift from pyrimidine to purine nucleoside phosphorylation.
|
| |
J Biol Chem,
281,
19273-19279.
|
 |
|
|
|
|
 |
M.Y.Mizutani,
Y.Takamatsu,
T.Ichinose,
K.Nakamura,
and
A.Itai
(2006).
Effective handling of induced-fit motion in flexible docking.
|
| |
Proteins,
63,
878-891.
|
 |
|
|
|
|
 |
G.Andrei,
J.Balzarini,
P.Fiten,
E.De Clercq,
G.Opdenakker,
and
R.Snoeck
(2005).
Characterization of herpes simplex virus type 1 thymidine kinase mutants selected under a single round of high-dose brivudin.
|
| |
J Virol,
79,
5863-5869.
|
 |
|
|
|
|
 |
H.Frederiksen,
D.Berenstein,
and
B.Munch-Petersen
(2004).
Effect of valine 106 on structure-function relation of cytosolic human thymidine kinase. Kinetic properties and oligomerization pattern of nine substitution mutants of V106.
|
| |
Eur J Biochem,
271,
2248-2256.
|
 |
|
|
|
|
 |
H.Merlitz,
T.Herges,
and
W.Wenzel
(2004).
Fluctuation analysis and accuracy of a large-scale in silico screen.
|
| |
J Comput Chem,
25,
1568-1575.
|
 |
|
|
|
|
 |
P.Schelling,
M.T.Claus,
R.Johner,
V.E.Marquez,
G.E.Schulz,
and
L.Scapozza
(2004).
Biochemical and structural characterization of (South)-methanocarbathymidine that specifically inhibits growth of herpes simplex virus type 1 thymidine kinase-transduced osteosarcoma cells.
|
| |
J Biol Chem,
279,
32832-32838.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Haouz,
V.Vanheusden,
H.Munier-Lehmann,
M.Froeyen,
P.Herdewijn,
S.Van Calenbergh,
and
M.Delarue
(2003).
Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
|
| |
J Biol Chem,
278,
4963-4971.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Monnerjahn,
and
M.Konrad
(2003).
Modulated nucleoside kinases as tools to improve the activation of therapeutic nucleoside analogues.
|
| |
Chembiochem,
4,
143-146.
|
 |
|
|
|
|
 |
G.Schuerman,
K.Van Hecke,
and
L.Van Meervelt
(2003).
Exploration of the influence of 5-iodo-2'-deoxyuridine incorporation on the structure of d[CACG(IDU)G].
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1525-1528.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.E.Bird,
J.Ren,
A.Wright,
K.D.Leslie,
B.Degrève,
J.Balzarini,
and
D.K.Stammers
(2003).
Crystal structure of varicella zoster virus thymidine kinase.
|
| |
J Biol Chem,
278,
24680-24687.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Ponomarev,
M.Doubrovin,
I.Serganova,
T.Beresten,
J.Vider,
A.Shavrin,
L.Ageyeva,
J.Balatoni,
R.Blasberg,
and
J.G.Tjuvajev
(2003).
Cytoplasmically retargeted HSV1-tk/GFP reporter gene mutants for optimization of noninvasive molecular-genetic imaging.
|
| |
Neoplasia,
5,
245-254.
|
 |
|
|
|
|
 |
C.Wurth,
U.Kessler,
J.Vogt,
G.E.Schulz,
G.Folkers,
and
L.Scapozza
(2001).
The effect of substrate binding on the conformation and structural stability of Herpes simplex virus type 1 thymidine kinase.
|
| |
Protein Sci,
10,
63-73.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.L.Gelpí,
S.G.Kalko,
X.Barril,
J.Cirera,
X.de La Cruz,
F.J.Luque,
and
M.Orozco
(2001).
Classical molecular interaction potentials: improved setup procedure in molecular dynamics simulations of proteins.
|
| |
Proteins,
45,
428-437.
|
 |
|
|
|
|
 |
J.Vogt,
R.Perozzo,
A.Pautsch,
A.Prota,
P.Schelling,
B.Pilger,
G.Folkers,
L.Scapozza,
and
G.E.Schulz
(2000).
Nucleoside binding site of herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography.
|
| |
Proteins,
41,
545-553.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Wouters,
and
P.Herdewijn
(1999).
5-Substituted pyrimidine 1,5-anhydrohexitols: conformational analysis and interaction with viral thymidine kinase.
|
| |
Bioorg Med Chem Lett,
9,
1563-1566.
|
 |
|
|
|
|
 |
M.Johansson,
A.R.van Rompay,
B.Degrève,
J.Balzarini,
and
A.Karlsson
(1999).
Cloning and characterization of the multisubstrate deoxyribonucleoside kinase of Drosophila melanogaster.
|
| |
J Biol Chem,
274,
23814-23819.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

