TRNA-pseudouridine synthase
tRNA-pseudouridine synthase II (TruB) from Thermotoga maritima catalyses the conversion of tRNA uridine to tRNA pseudouridine at position 55 in tRNA. The formation of pseudouridine is shown to be important for the structural integrity of tRNA.
TruB is responsible for the pseudouridine residue present in the T loops of virtually all tRNAs. TruB recognises the preformed 3-D structure of the T loop primarily through shape complementarity. It accesses its substrate uridyl residue by flipping out the nucleotide and disrupts the tertiary structure of tRNA [PMID:11779468].
The catalytic domain consists of two subdomains, each of which has an alpha+beta structure that has some similarity to the ferredoxin-like fold (note: some pseudouridine synthases contain additional domains). The active site is the most conserved structural region of the superfamily and is located between the two homologous domains. There are four distinct families of pseudouridine synthases that share no global sequence similarity, but which do share the same fold of their catalytic domain(s) and uracil-binding site and are descended from a common molecular ancestor. [PMID:10529181]
There is still much debate as to the exact mechanism of this enzyme.
Reference Protein and Structure
- Sequence
-
Q9WZW0
 (5.4.99.25)
(5.4.99.25)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Thermotoga maritima MSB8 (Bacteria)

- PDB
-
1ze1
- Conformational Change of Pseudouridine 55 Synthase upon Its Association with RNA Substrate
(2.9 Å)



- Catalytic CATH Domains
-
3.30.2350.10
 (see all for 1ze1)
(see all for 1ze1)
- Cofactors
- Water (1)
Enzyme Reaction (EC:5.4.99.25)
Enzyme Mechanism
Introduction
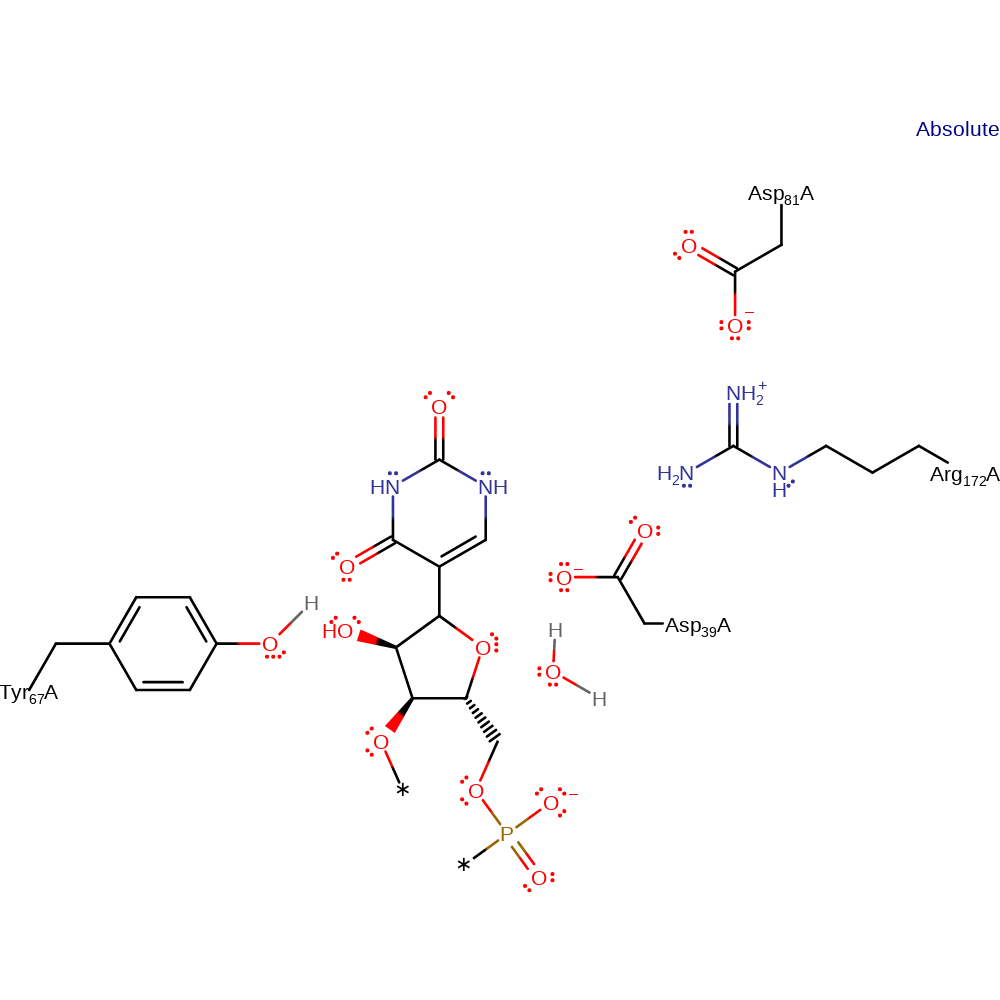
This entry represents the so-called "glycal mechanism." This mechanism begins with deprotonation of C2′ to eliminate the pyrimidine ring and form the glycal intermediate followed by reattachment of the repositioned pyrimidine ring to form the C-glycoside. In this proposal, Asp39 acts as a general acid/base, rather than a catalytic nucleophile.
Catalytic Residues Roles
| UniProt | PDB* (1ze1) | ||
| Asp39 | Asp39A | Acts as a general acid/base. | proton acceptor, proton donor |
| Tyr67 | Tyr67A | Exact role in this mechanism is unclear, but likely to be involved in the stabilisation of the intermediates. | electrostatic stabiliser |
| Asp81, Arg172 | Asp81A, Arg172A | Essential for catalytic activity, has no effect on binding. Thought to be essential to modulate the reactivity of the nucleophilic aspartate residue. | increase basicity, increase acidity |
Chemical Components
intramolecular elimination, proton transfer, assisted tautomerisation (not keto-enol), bimolecular nucleophilic addition, inferred reaction step, native state of enzyme regeneratedReferences
- Veerareddygari GR et al. (2016), J Am Chem Soc, 138, 7852-7855. The Pseudouridine Synthases Proceed through a Glycal Intermediate. DOI:10.1021/jacs.6b04491. PMID:27292228.
- Spenkuch F et al. (2014), RNA Biol, 11, 1540-1554. Pseudouridine: Still mysterious, but never a fake (uridine)! DOI:10.4161/15476286.2014.992278. PMID:25616362.
- Friedt J et al. (2014), Nucleic Acids Res, 42, 3857-3870. An arginine-aspartate network in the active site of bacterial TruB is critical for catalyzing pseudouridine formation. DOI:10.1093/nar/gkt1331. PMID:24371284.
- Phannachet K et al. (2005), Biochemistry, 44, 15488-15494. Dissecting the Roles of a Strictly Conserved Tyrosine in Substrate Recognition and Catalysis by Pseudouridine 55 Synthase†. DOI:10.1021/bi050961w. PMID:16300397.
- Hamilton CS et al. (2005), Arch Biochem Biophys, 433, 322-334. The roles of the essential Asp-48 and highly conserved His-43 elucidated by the pH dependence of the pseudouridine synthase TruB. DOI:10.1016/j.abb.2004.09.009. PMID:15581587.
- Hoang C et al. (2001), Cell, 107, 929-939. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. PMID:11779468.
- Huang L et al. (1998), Biochemistry, 37, 344-351. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. DOI:10.1021/bi971874. PMID:9425056.
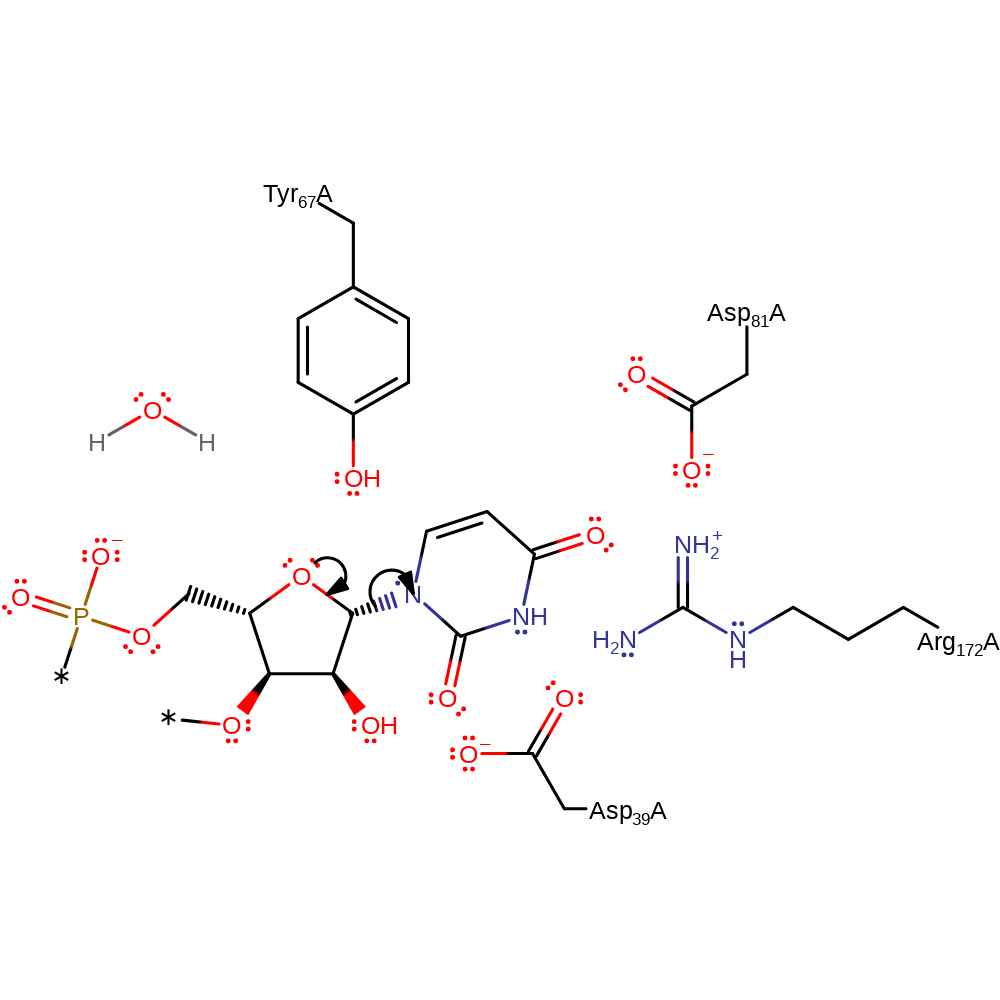
Step 1. Intramolecular elimination of the uridine group to produce an oxycarbenium intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr67A | electrostatic stabiliser |
Chemical Components
ingold: intramolecular eliminationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp81A | increase basicity |
| Arg172A | increase basicity |
| Tyr67A | electrostatic stabiliser |
| Asp39A | proton acceptor |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol)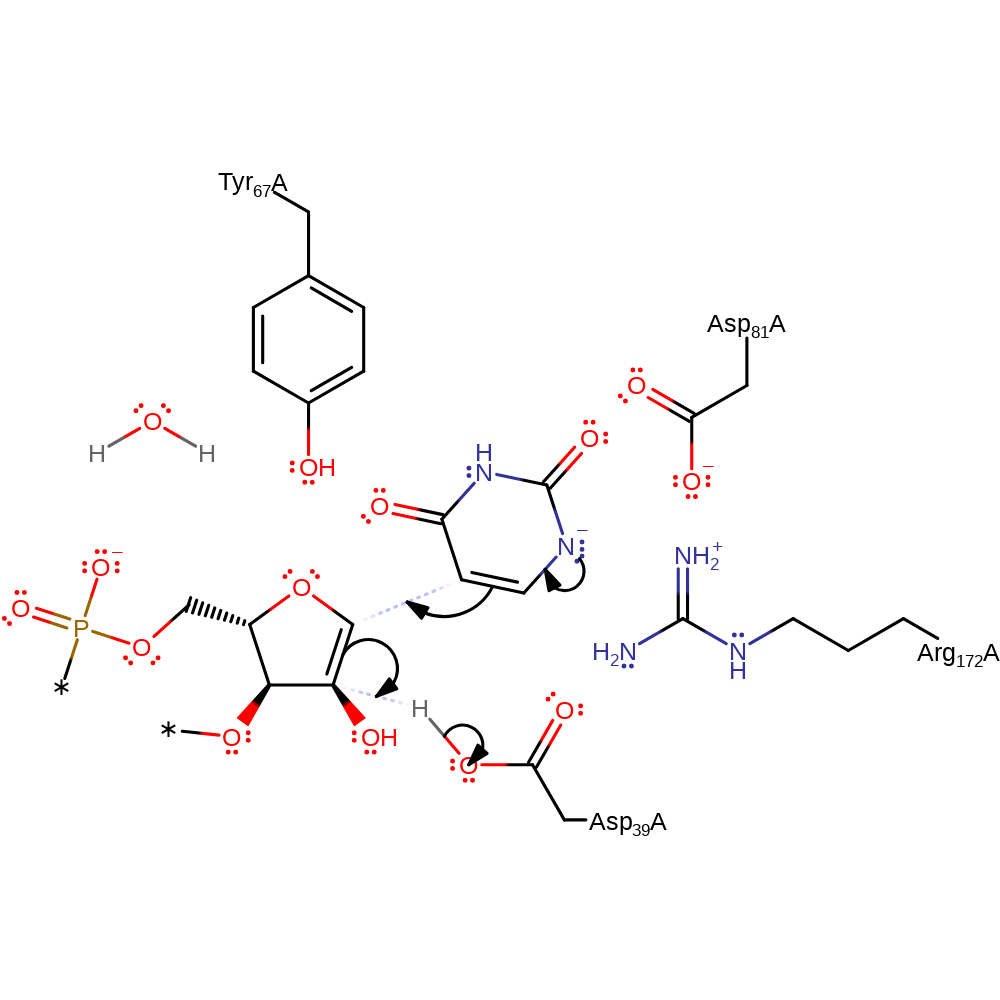
Step 3. The pyrimidine base rotates in the active site. The negative charge on the nitrogen then initiates a nucleophilic attack onto the ribose intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr67A | electrostatic stabiliser |
| Asp81A | increase acidity |
| Arg172A | increase acidity |
| Asp39A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition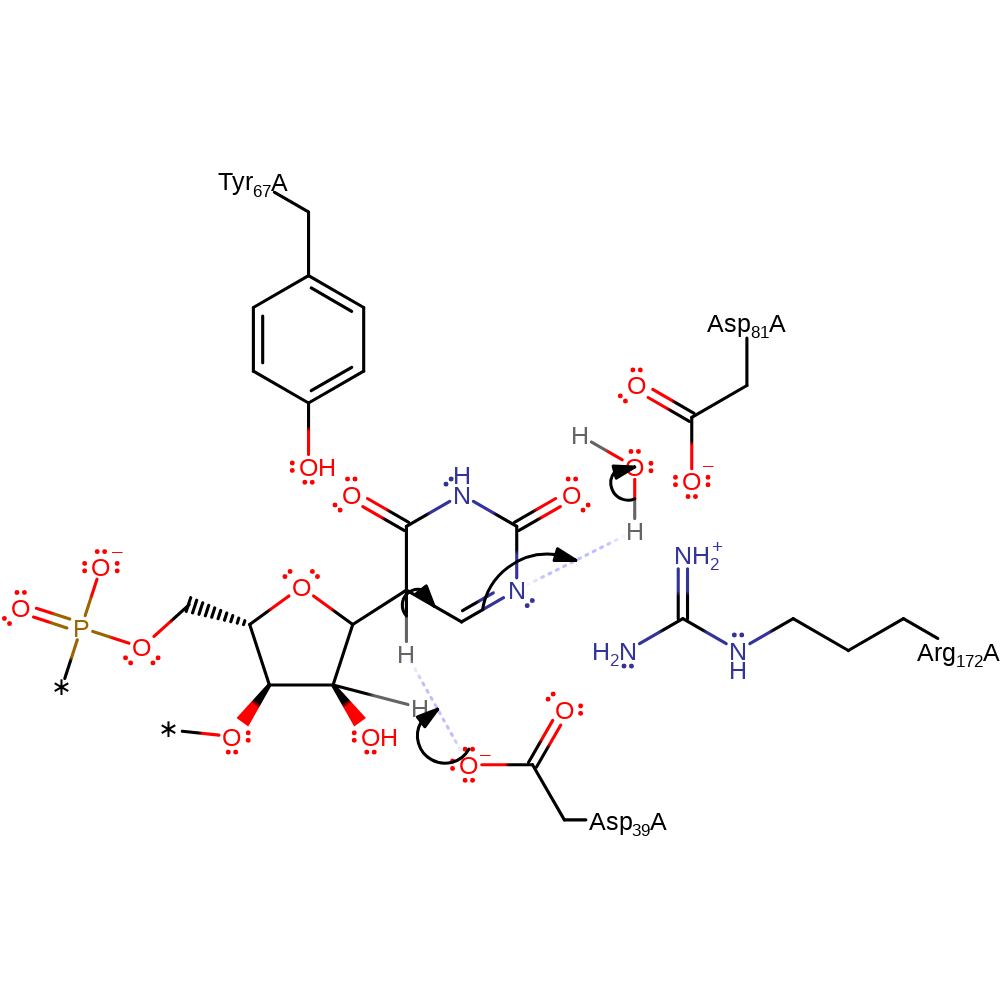
Step 4. Asp39 abstracts a proton from the tetrahedral carbon intermediate, generating the pseudouridine product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp81A | increase basicity |
| Arg172A | increase basicity |
| Tyr67A | electrostatic stabiliser |
| Asp39A | proton acceptor |
Chemical Components
assisted tautomerisation (not keto-enol), proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp81A | increase acidity |
| Arg172A | increase acidity |
| Asp39A | proton donor |
Chemical Components
proton transfer, inferred reaction step, native state of enzyme regeneratedIntroduction
This mechanism represents the acyl-enzyme proposal:
- Asp 39 nucleophilically attacks the C1' atom of the ribose, detaching the uracil base from the ribose.
- Rotation of the detached base occurs, and re-attachment of the rotated base results in the formation of the C5-C1' bond between the base and the ribose, and detaching Asp 39.
- The OH group of Tyr 67 donates its proton to N1 atom of the base, while abstracting the C5 proton from the base.
Catalytic Residues Roles
| UniProt | PDB* (1ze1) | ||
| Asp39 | Asp39A | Acts as a nucleophile by attacking C1' of the scissile bond. | nucleofuge, nucleophile |
| Tyr67 | Tyr67A | The negative pi-cloud of Tyr 67 stabilises the oxocarbenium intermediate. Tyr 67 also donates its proton to N1 of the base, while abstracting the C5 proton. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay, van der waals interaction, electrostatic stabiliser |
| Asp81, Arg172 | Asp81A, Arg172A | Essential for catalytic activity, has no effect on binding. Thought to be essential to modulate the reactivity of the nucleophilic aspartate residue. | increase electrophilicity, activator |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, intermediate formation, enzyme-substrate complex cleavage, assisted tautomerisation (not keto-enol), proton transfer, proton relay, overall product formed, intermediate terminated, native state of enzyme regeneratedReferences
- Mueller EG (2002), Nat Struct Biol, 9, 320-322. Chips off the old block. DOI:10.1038/nsb0502-320. PMID:11976723.
- Friedt J et al. (2014), Nucleic Acids Res, 42, 3857-3870. An arginine-aspartate network in the active site of bacterial TruB is critical for catalyzing pseudouridine formation. DOI:10.1093/nar/gkt1331. PMID:24371284.
- Spenkuch F et al. (2014), RNA Biol, 11, 1540-1554. Pseudouridine: Still mysterious, but never a fake (uridine)! DOI:10.4161/15476286.2014.992278. PMID:25616362.
- Miracco EJ et al. (2011), J Am Chem Soc, 133, 11826-11829. The Products of 5-Fluorouridine by the Action of the Pseudouridine Synthase TruB Disfavor One Mechanism and Suggest Another. DOI:10.1021/ja201179f. PMID:21744792.
- Phannachet K et al. (2005), Biochemistry, 44, 15488-15494. Dissecting the Roles of a Strictly Conserved Tyrosine in Substrate Recognition and Catalysis by Pseudouridine 55 Synthase†. DOI:10.1021/bi050961w. PMID:16300397.
- Hamilton CS et al. (2005), Arch Biochem Biophys, 433, 322-334. The roles of the essential Asp-48 and highly conserved His-43 elucidated by the pH dependence of the pseudouridine synthase TruB. DOI:10.1016/j.abb.2004.09.009. PMID:15581587.
- Hoang C et al. (2005), Protein Sci, 14, 2201-2206. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. DOI:10.1110/ps.051493605. PMID:15987897.
- Phannachet K et al. (2004), Nucleic Acids Res, 32, 1422-1429. Conformational change of pseudouridine 55 synthase upon its association with RNA substrate. DOI:10.1093/nar/gkh287. PMID:14990747.
- Chaudhuri BN et al. (2004), J Biol Chem, 279, 24585-24591. Crystal Structure of the Apo Forms of 55 tRNA Pseudouridine Synthase from Mycobacterium tuberculosis: A HINGE AT THE BASE OF THE CATALYTIC CLEFT. DOI:10.1074/jbc.m401045200. PMID:15028724.
- Hoang C et al. (2001), Cell, 107, 929-939. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. PMID:11779468.
- Foster PG et al. (2000), Nat Struct Biol, 7, 23-27. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. DOI:10.1038/71219. PMID:10625422.
- Huang L et al. (1998), Biochemistry, 37, 344-351. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. DOI:10.1021/bi971874. PMID:9425056.
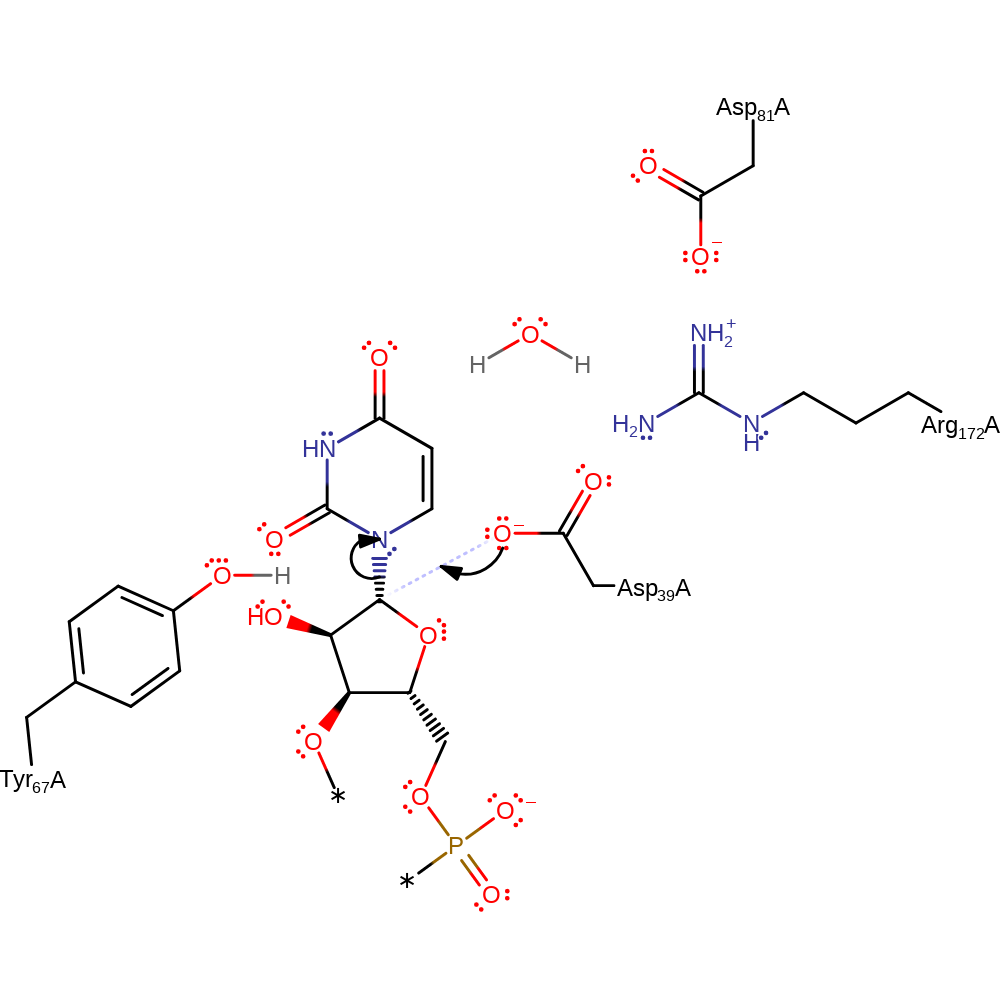
Step 1. Asp39 initiates a nucleophilic attack upon the C1 of the uridine tRNA in a substitution reaction, displacing the uridine base as an anion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp81A | activator |
| Arg172A | activator |
| Asp39A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, intermediate formation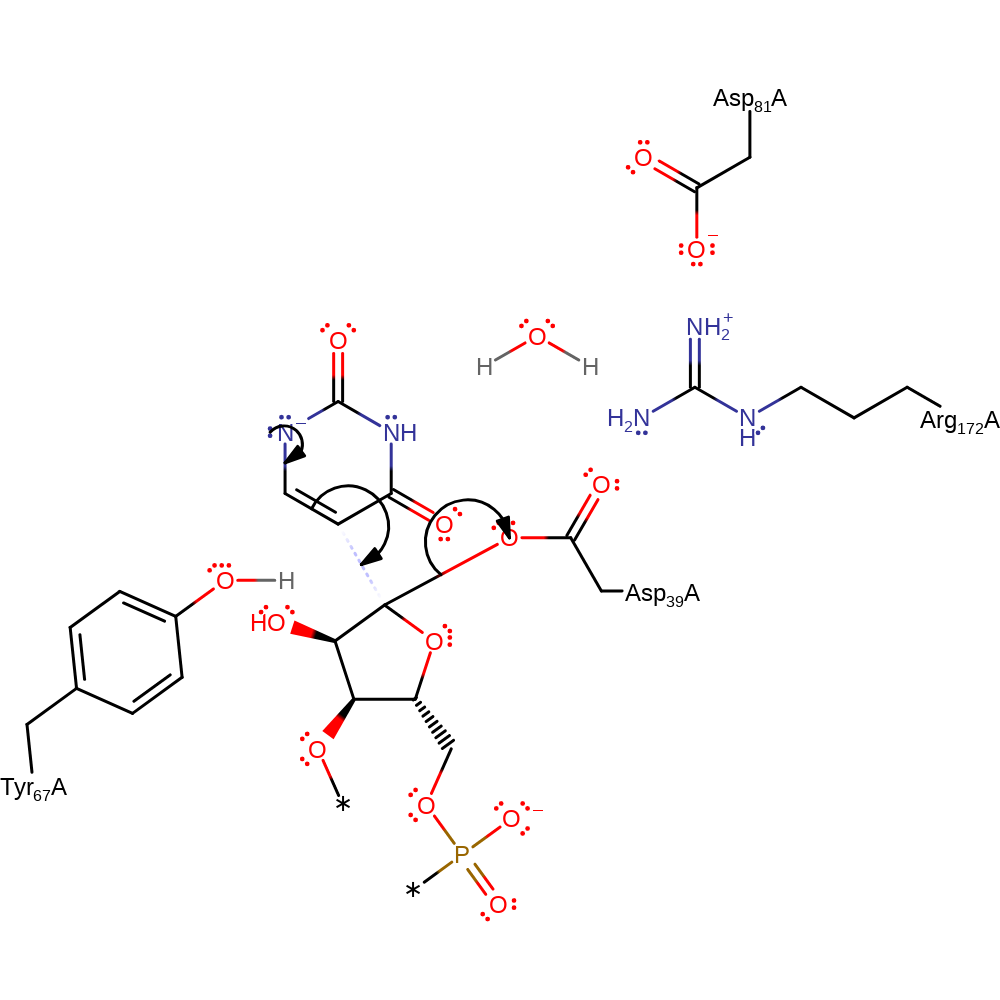
Step 2. The uridine undergoes rotation within the active site. The anion then initiates double bond rearrangement, which results in nucleophilic attack of the transient carbanion (from the C=C) on the covalently bound tRNA molecule in a substitution reaction, displacing Asp39.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr67A | electrostatic stabiliser, hydrogen bond donor, van der waals interaction |
| Asp81A | increase electrophilicity |
| Arg172A | increase electrophilicity |
| Asp39A | nucleofuge |
Chemical Components
ingold: bimolecular nucleophilic substitution, enzyme-substrate complex cleavage, intermediate formation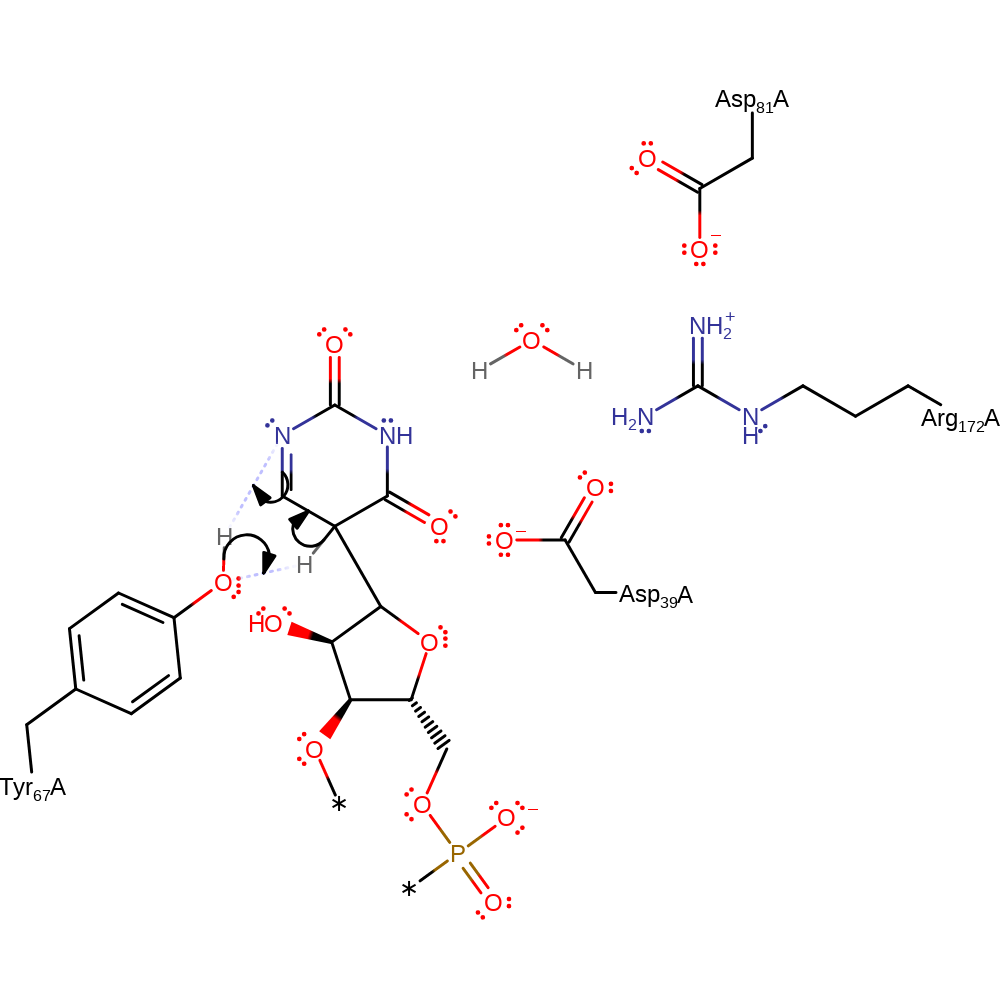
Step 3. The pseudouridine then deprotonates Tyr67, which in turn deprotonates the sugar bound carbon of the ring forming the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr67A | proton relay, hydrogen bond acceptor, hydrogen bond donor, van der waals interaction, proton donor, proton acceptor |
Chemical Components
assisted tautomerisation (not keto-enol), proton transfer, proton relay, overall product formed, intermediate terminated, native state of enzyme regeneratedIntroduction
In this proposal, a Michael Adduct is formed through the nucleophilic attack at C6 of the target uridine by Asp 39 and leads to cleavage of the N1-C1' glycosidic bond. A 180 degree rotation of the uracil ring about the new C6−Odelta1 bond juxtaposes C5 with C1' and allows for carbon-carbon formation, followed by breakdown of the covalent PsiS I−tRNA.
Catalytic Residues Roles
| UniProt | PDB* (1ze1) | ||
| Asp39 | Asp39A | Acts as a catalytic nucleophile. | covalently attached, nucleofuge, nucleophile |
| Tyr67 | Tyr67A | Acts as a general acid/base. | proton acceptor, proton donor |
| Asp81, Arg172 | Asp81A, Arg172A | Essential for catalytic activity, has no effect on binding. Thought to be essential to modulate the reactivity of the nucleophilic aspartate residue. | increase electrophilicity, activator |
Chemical Components
enzyme-substrate complex formation, bimolecular nucleophilic addition, proton transfer, assisted keto-enol tautomerisation, unimolecular homolytic elimination, bimolecular eliminationReferences
- Mueller EG (2002), Nat Struct Biol, 9, 320-322. Chips off the old block. DOI:10.1038/nsb0502-320. PMID:11976723.
- Friedt J et al. (2014), Nucleic Acids Res, 42, 3857-3870. An arginine-aspartate network in the active site of bacterial TruB is critical for catalyzing pseudouridine formation. DOI:10.1093/nar/gkt1331. PMID:24371284.
- Spenkuch F et al. (2014), RNA Biol, 11, 1540-1554. Pseudouridine: Still mysterious, but never a fake (uridine)! DOI:10.4161/15476286.2014.992278. PMID:25616362.
- Hamilton CS et al. (2006), Biochemistry, 45, 12029-12038. Mechanistic Investigations of the Pseudouridine Synthase RluA Using RNA Containing 5-Fluorouridine†. DOI:10.1021/bi061293x. PMID:17002302.
- Hamilton CS et al. (2005), Arch Biochem Biophys, 433, 322-334. The roles of the essential Asp-48 and highly conserved His-43 elucidated by the pH dependence of the pseudouridine synthase TruB. DOI:10.1016/j.abb.2004.09.009. PMID:15581587.
- Hoang C et al. (2005), Protein Sci, 14, 2201-2206. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. DOI:10.1110/ps.051493605. PMID:15987897.
- Phannachet K et al. (2004), Nucleic Acids Res, 32, 1422-1429. Conformational change of pseudouridine 55 synthase upon its association with RNA substrate. DOI:10.1093/nar/gkh287. PMID:14990747.
- Chaudhuri BN et al. (2004), J Biol Chem, 279, 24585-24591. Crystal Structure of the Apo Forms of 55 tRNA Pseudouridine Synthase from Mycobacterium tuberculosis: A HINGE AT THE BASE OF THE CATALYTIC CLEFT. DOI:10.1074/jbc.m401045200. PMID:15028724.
- Pan H et al. (2003), Proc Natl Acad Sci U S A, 100, 12648-12653. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. DOI:10.1073/pnas.2135585100. PMID:14566049.
- Hoang C et al. (2001), Cell, 107, 929-939. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. PMID:11779468.
- Foster PG et al. (2000), Nat Struct Biol, 7, 23-27. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. DOI:10.1038/71219. PMID:10625422.
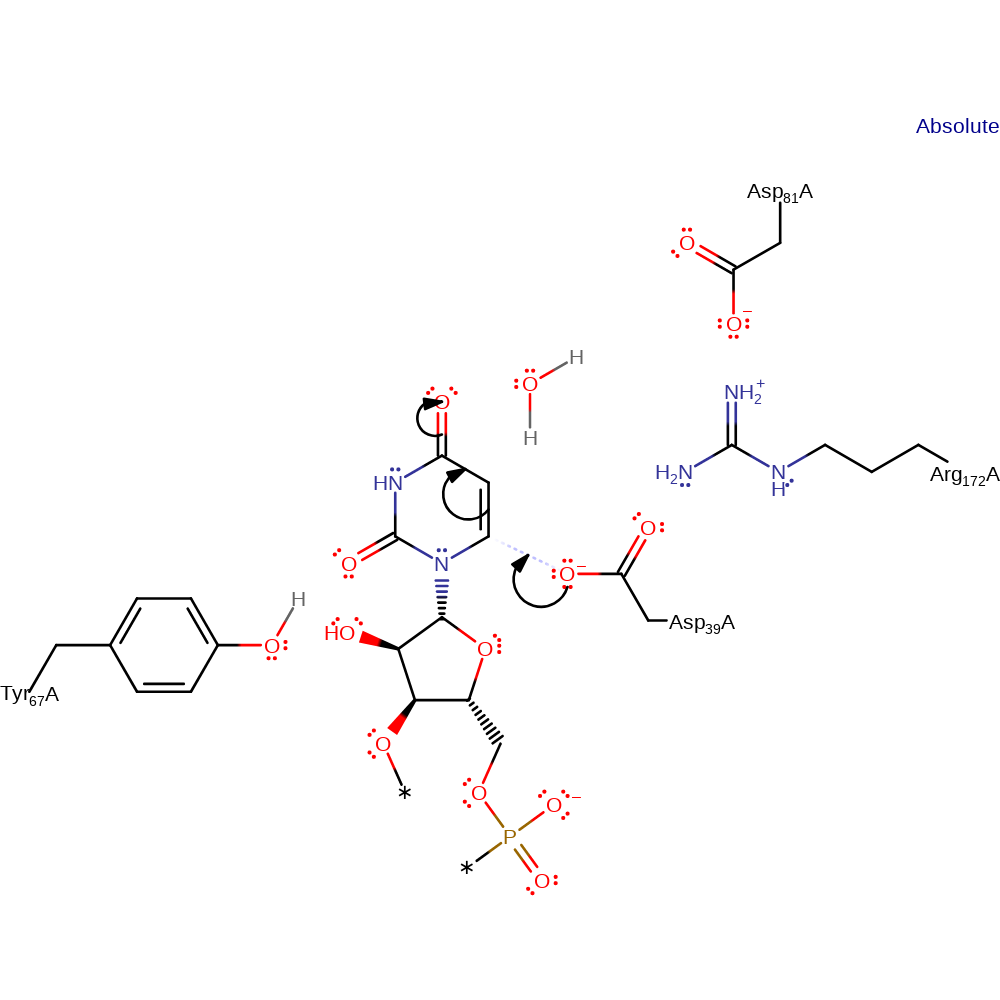
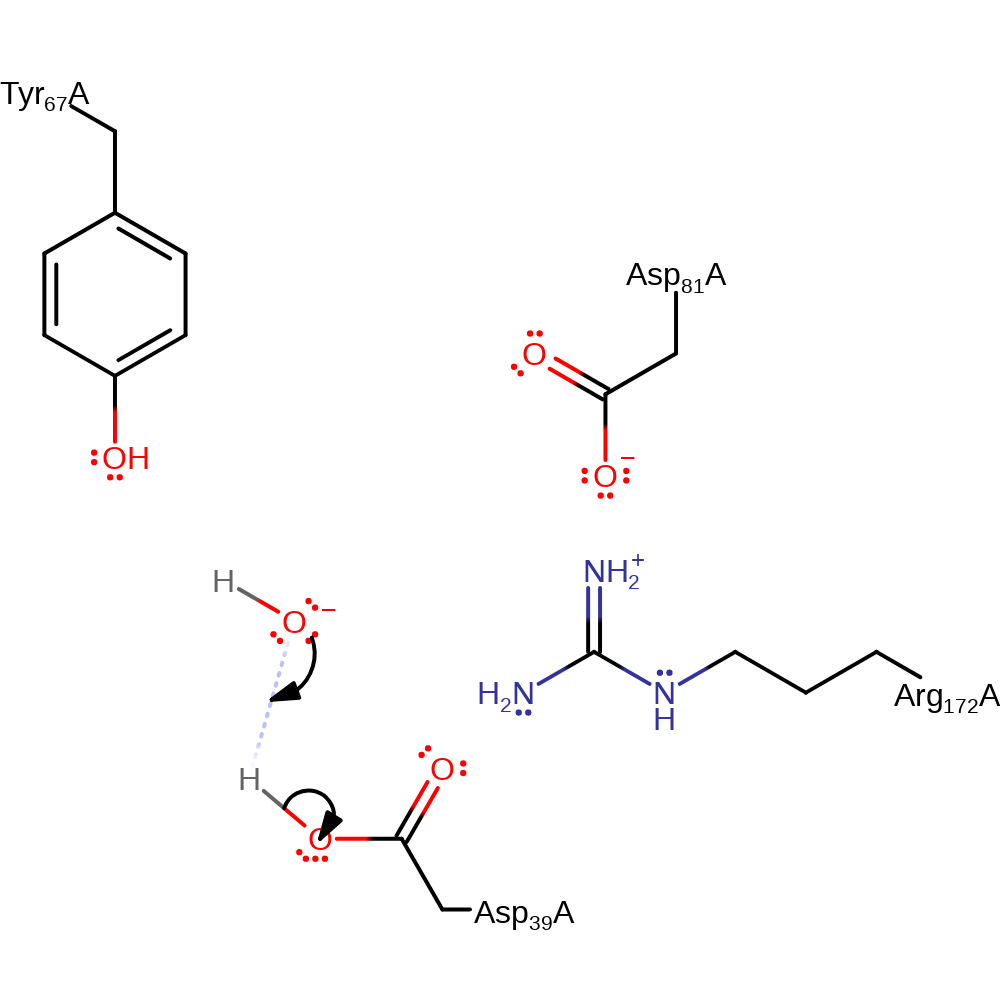
Step 1. Asp39 initiates a nucleophilic attack upon the C1 of the uridine tRNA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp81A | activator |
| Arg172A | activator |
| Asp39A | nucleophile |
Chemical Components
enzyme-substrate complex formation, ingold: bimolecular nucleophilic addition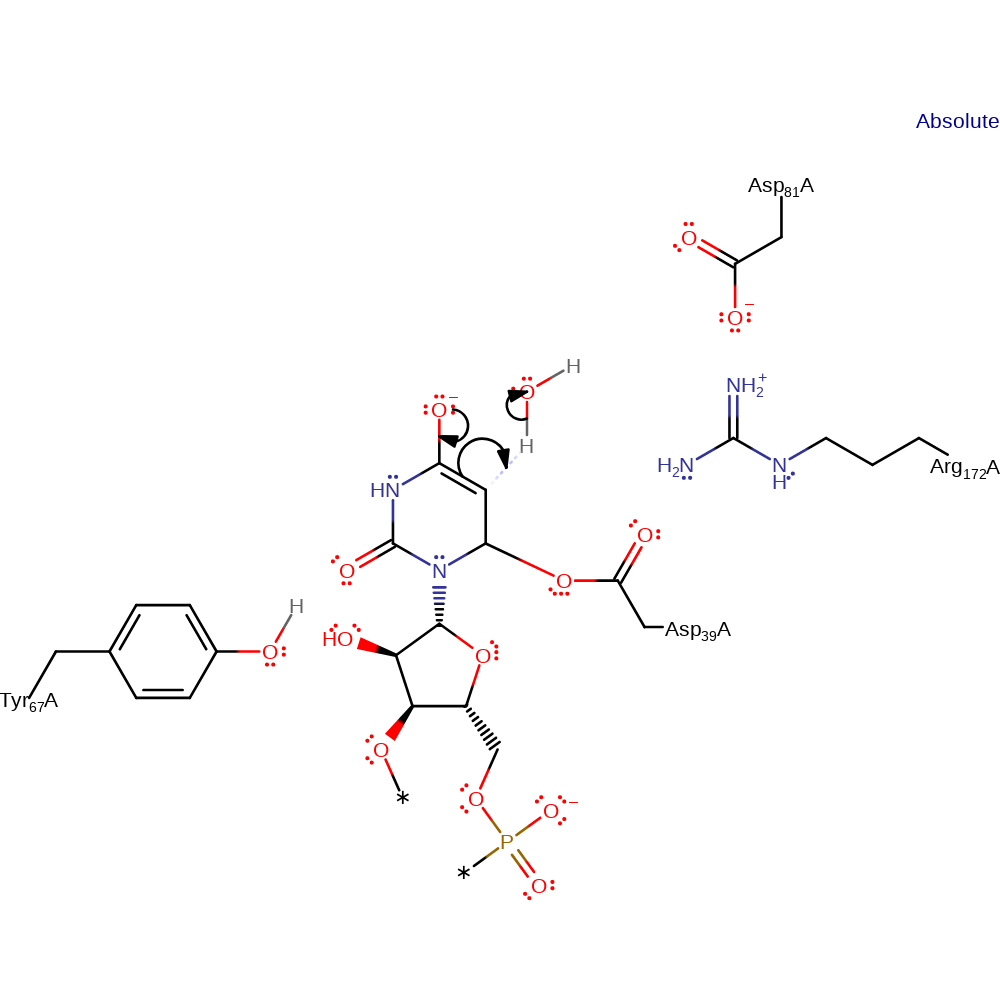
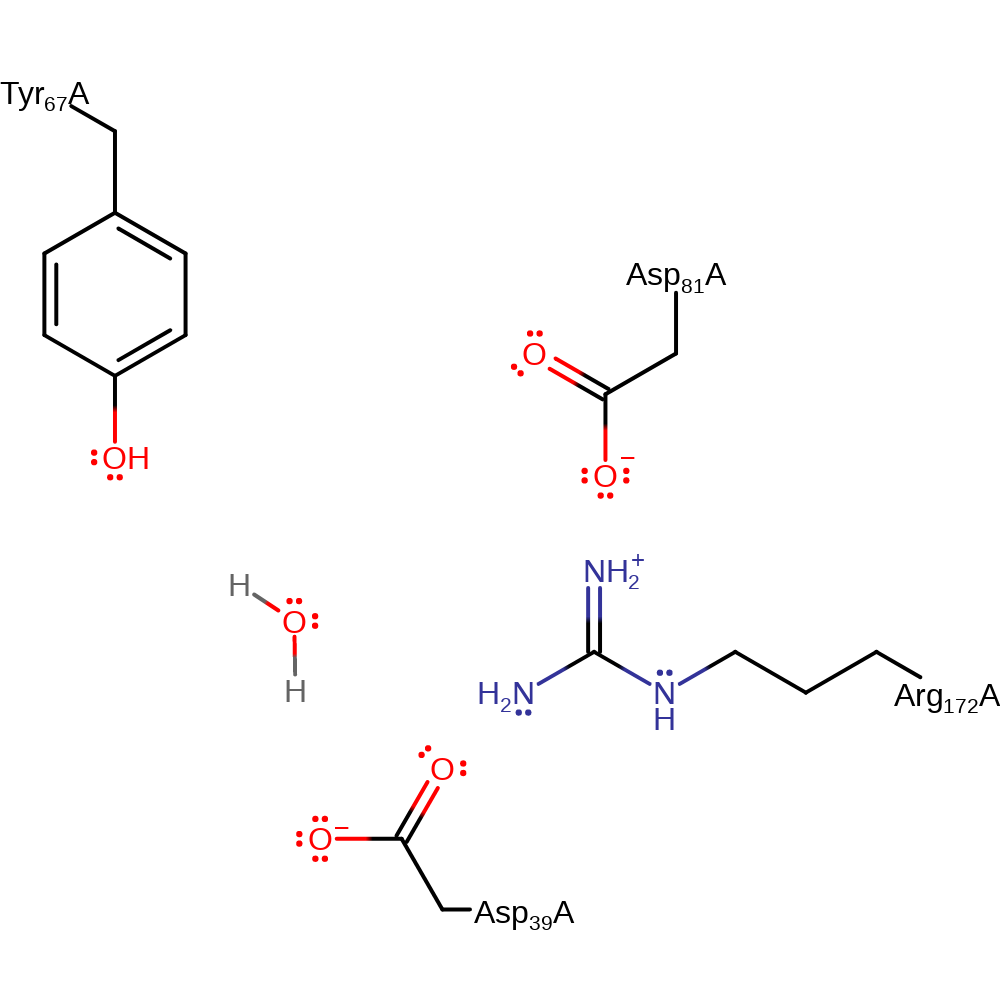
Step 2. The oxyanion intermediate collapses, deprotonating an unidentified base (shown as water here for mechanism purposes).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp39A | covalently attached |
Chemical Components
proton transfer, assisted keto-enol tautomerisationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp39A | covalently attached |
Chemical Components
ingold: unimolecular homolytic elimination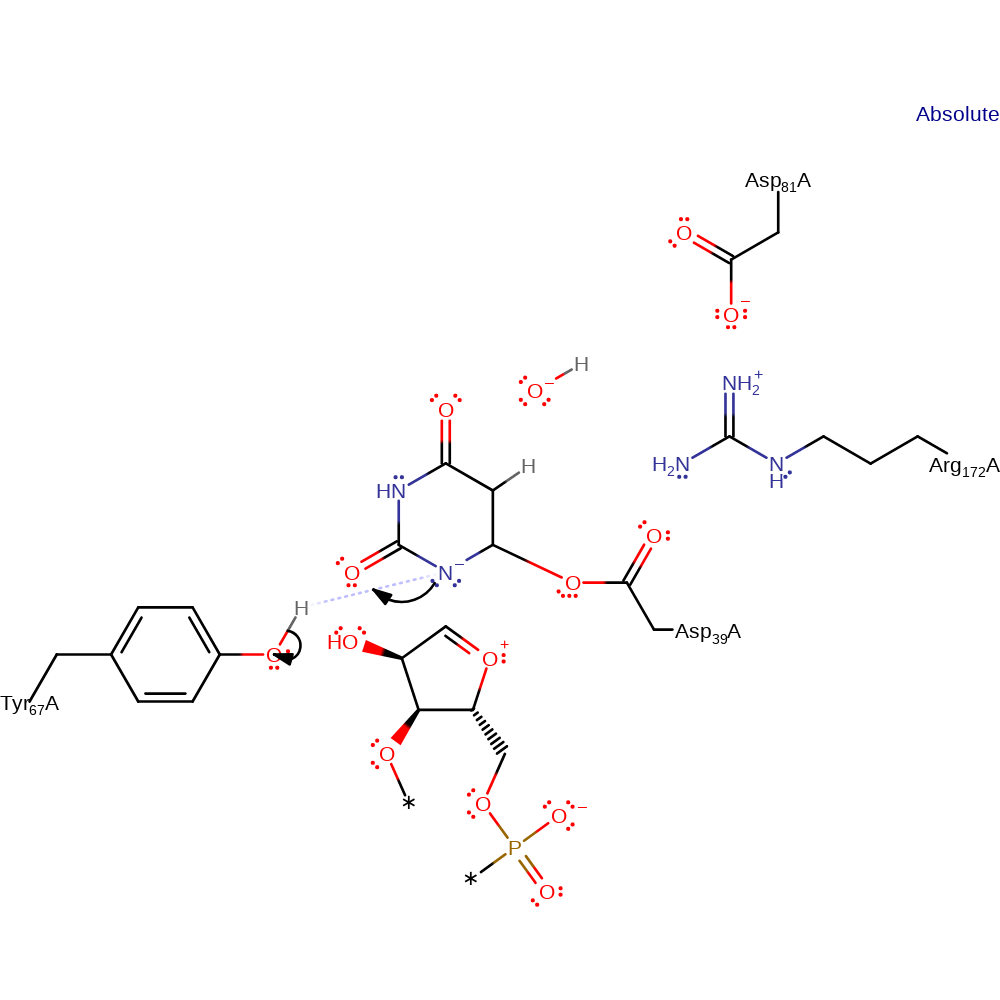
Step 4. The negatively charged N of the uridine base abstracts a proton from a general base (shown here as Tyr67).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp39A | covalently attached |
| Tyr67A | proton donor |
Chemical Components
proton transfer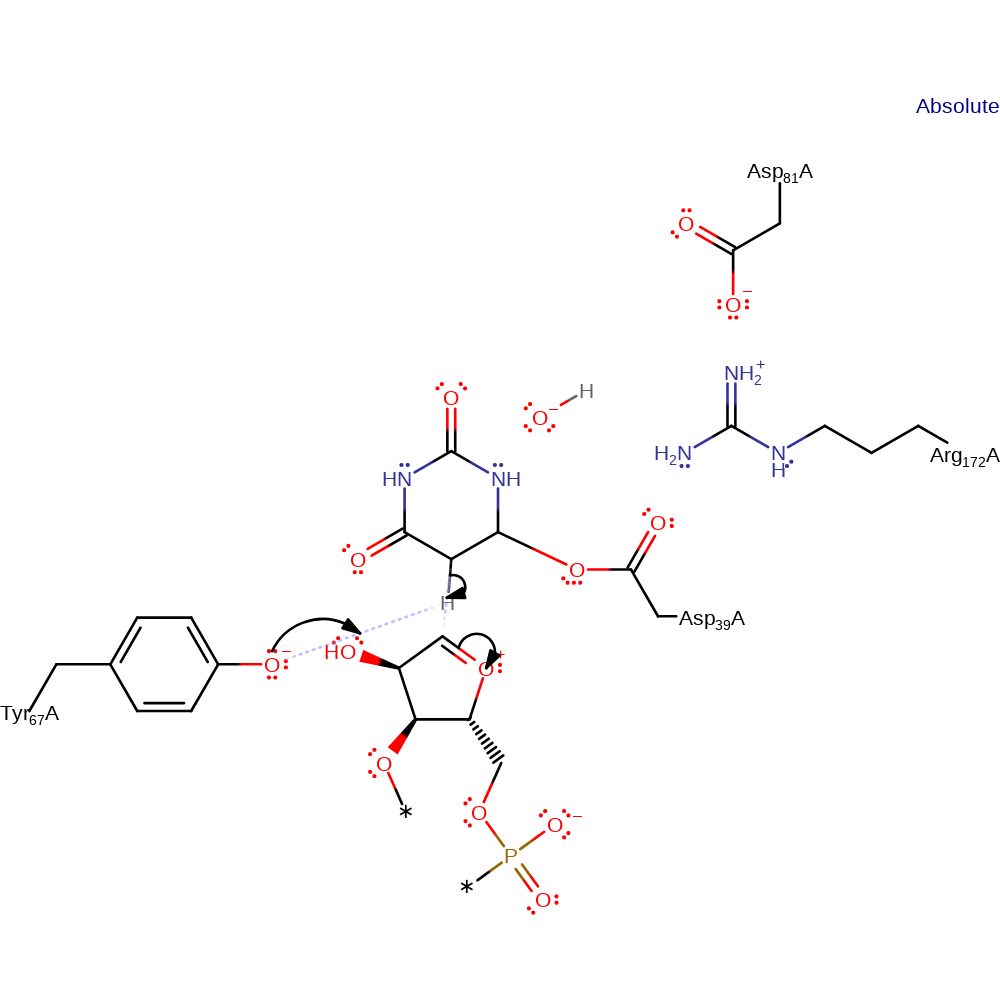
Step 5. The Uridine rotates in the active site. The base (Tyr69) deprotonates which causes the uridine to attack the sugar ring.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp39A | covalently attached |
| Tyr67A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition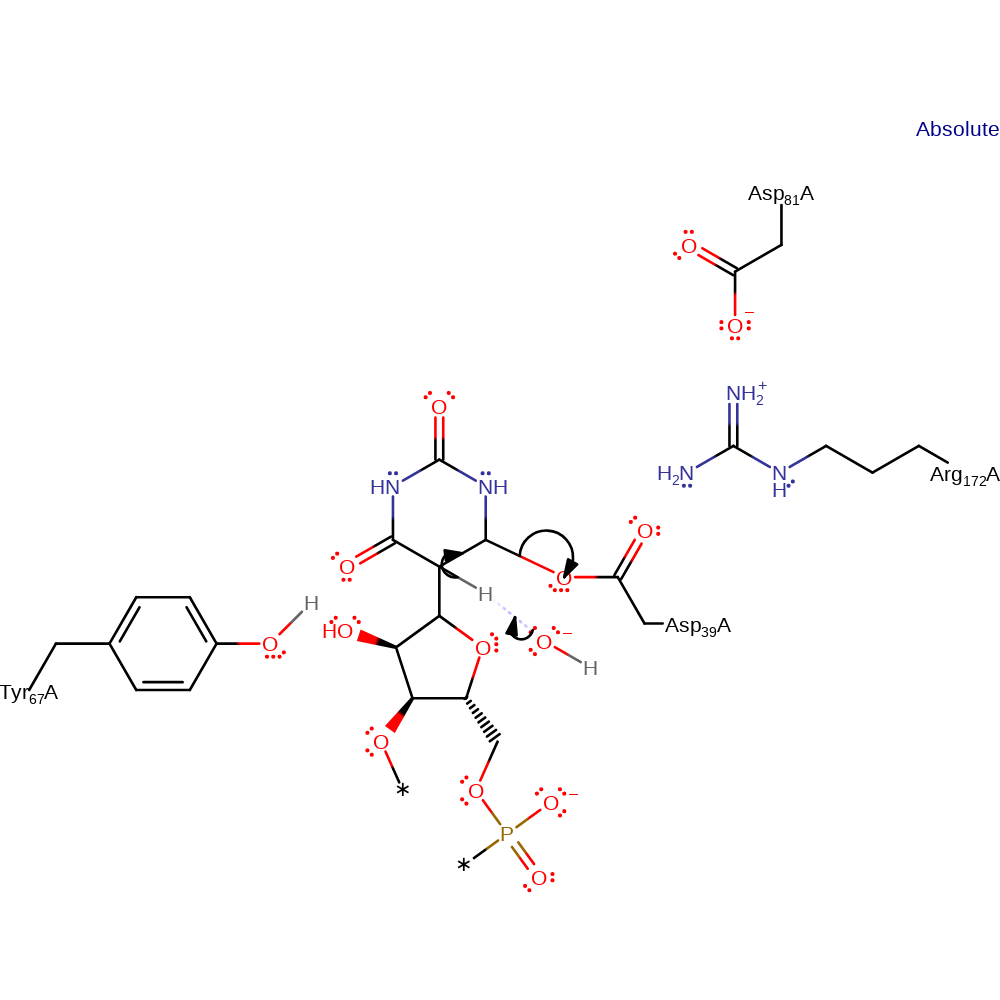
Step 6. An unidentified base (shown as water here) deprotonates the carbon newly attached to the sugar ring, eliminating the Asp and generating the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp81A | increase electrophilicity |
| Arg172A | increase electrophilicity |
| Asp39A | nucleofuge |


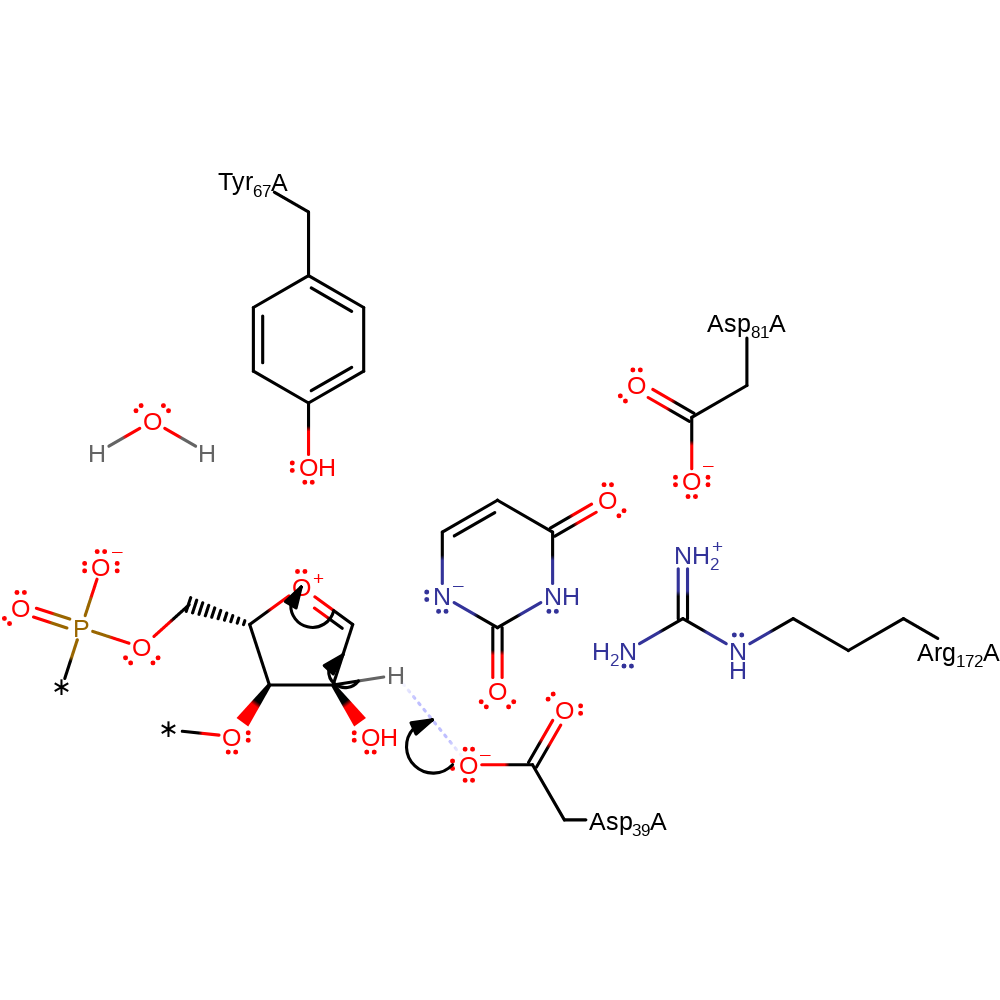
 Download:
Download: 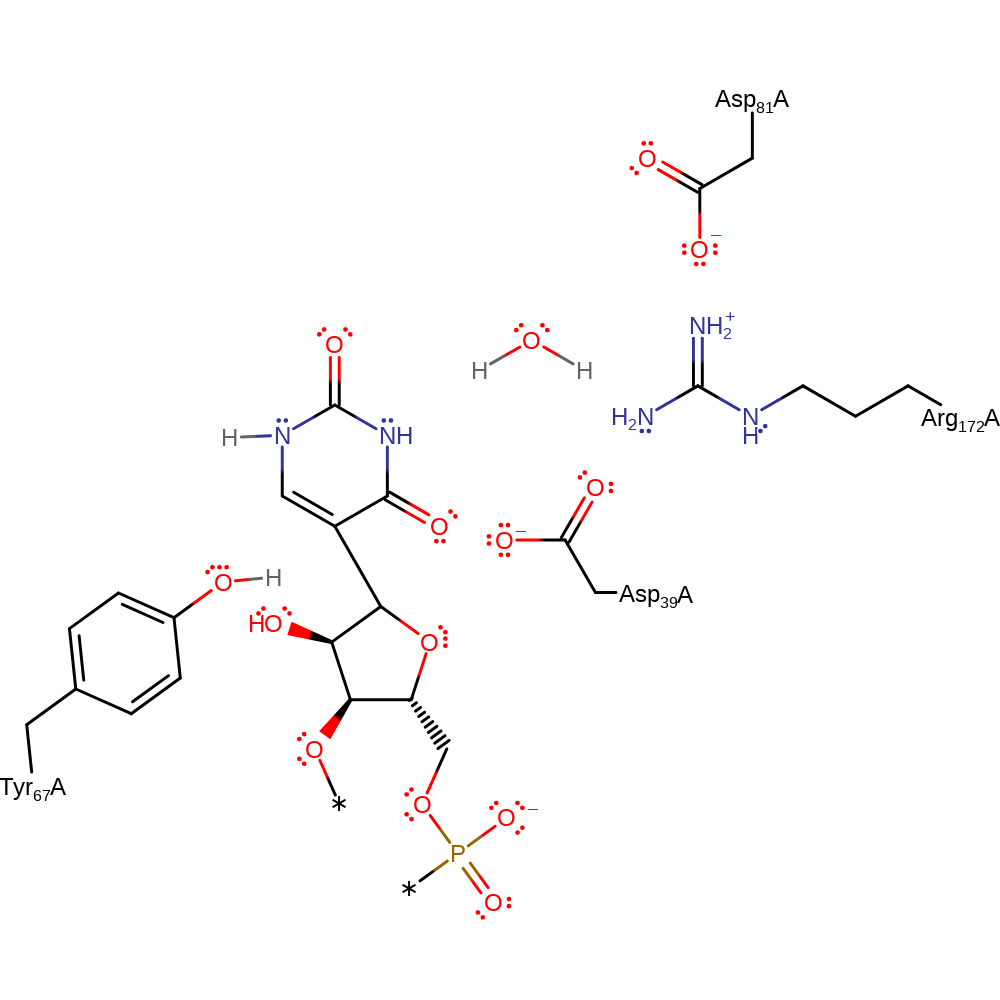 Download:
Download: 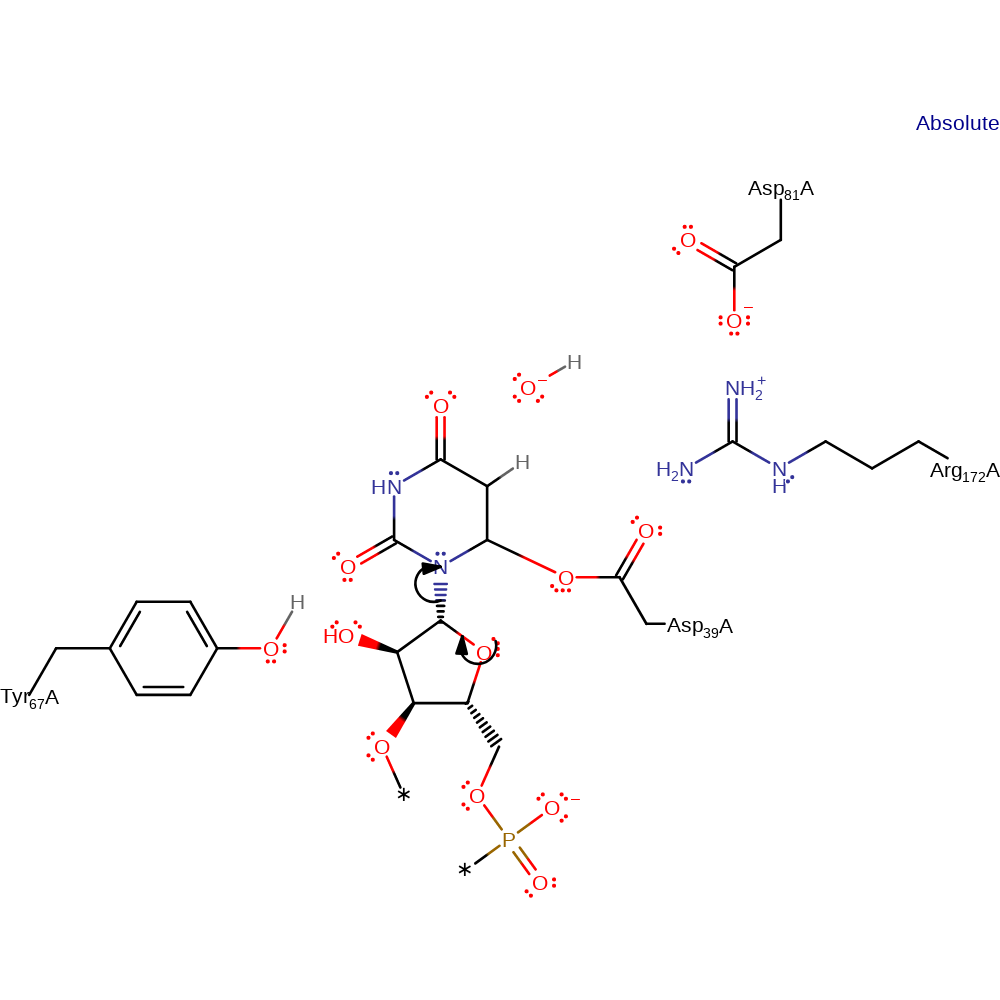
 Download:
Download: