Phenol 2-monooxygenase
Phenol 2-monooxygenase, also known as phenol hydroxylase (PHHY) belongs to a class of flavin enzymes commonly referred to as the aromatic hydroxylases. It catalyses the conversion of simple phenols to their o-diol derivatives. The enzyme is a homodimer where each monomer of 76kDa contains noncovalently bound flavin adenine dinucleotide (FAD) The enzyme can hydroxylate simple, amino, methyl, hydroxy and halogen phenols, although it does not show any activity towards phenols carrying carboxyl groups on the benzene nucleus or its side chain.
Next to carbohydrates, phenolic compounds are the second most abundant group of natural products, many are also being produced by industry, with many of these being resistant to degradation. The reaction catalysed by phenol hydroxylase are of particular interest for industrial waste detoxification.
Reference Protein and Structure
- Sequence
-
P15245
 (1.14.13.7)
(1.14.13.7)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Cutaneotrichosporon cutaneum (Yeast)

- PDB
-
1foh
- PHENOL HYDROXYLASE FROM TRICHOSPORON CUTANEUM
(2.4 Å)



- Catalytic CATH Domains
-
3.30.9.10
 3.50.50.60
3.50.50.60  (see all for 1foh)
(see all for 1foh)
- Cofactors
- Fadh2(2-) (1)
Enzyme Reaction (EC:1.14.13.7)
Enzyme Mechanism
Introduction
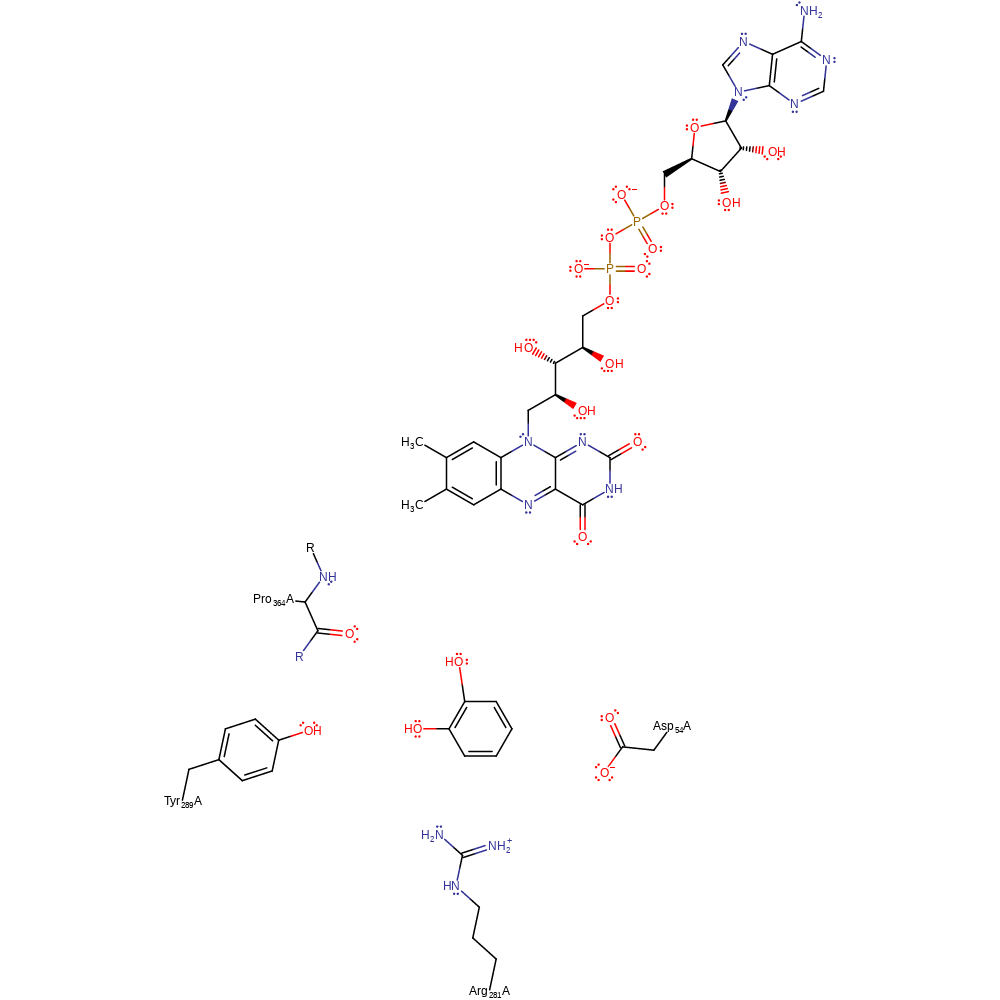
It has been propose that a movement of FAD takes place in concert with a large conformational change of residues 170-210 during catalysis. The catalytic mechanism of PHHY is complex, involving FAD and three substrates: molecular oxygen, phenol and NADPH. The overall mechanism is of the type bi-uni-uni-bi ping pong. While in the past the mechanism has been compared to p-hydroxy-benzoate hydroxylase, the enzymes are now though to operate differently.
The flavin cofactor is first reduced by NADPH. After releasing NADP+, the reduced flavin reacts with oxygen to form C4a-hydroperoxyflavin that it used to hydroxylate the substrate, phenol, or its derivatives at the ortho position. The immediate product undergoes further change to form C4a-hydroxyflavin. The enzyme finishes the reaction cycle by releasing a molecule of water to return the flavin to the oxidised state.
Phenol is suitably placed for attack at the ortho position through a hydrogen bond to Tyr289. This residue is also thought to control FAD conformation, and therefore direct FAD reduction. Modelling suggests that Tyr289 is also able to form a hydrogen bond to the oxygen of C4a-peroxoflavin intermediate. Asp54 together with Arg281forms a hydrogen bond to the phenolic oxygen, thereby increasing its partial negative charge, the side chain could also stabilise the positive charge on the substrate developed in the transition state. However, when these residue are mutated, their mutant forms do not decrease catalytic activity dramatically. The one residue which has been implicated in catalysis by mutagenesis, Pro364, is thought to use its carbonyl backbone to hydrogen bond to phenol substrate and so direct nucleophilic attack of the hydroperoxide at C2.
Catalytic Residues Roles
| UniProt | PDB* (1foh) | ||
| Pro365 (main-N) | Pro364A (main-N) | The residue's backbone carbonyl is implicated in directing nucleophilic attack of the phenol C2 at C4a- hydroperoxyflavin. | activator, electrostatic stabiliser |
| Arg282 | Arg281A | The residue is thought to act with Asp54 within the active site to polarise the phenolic OH bond through hydrogen bonding interactions, activating the oxygen towards conjugate attack at the C4a- hydroperoxyflavin intermediate. These residues, however, do not fulfil the role of a general base towards the phenol substrate. | electrostatic stabiliser |
| Tyr290 | Tyr289A | The residue affects the equilibrium between the two conformations of FAD and is thought to help the flavin move into a conformation necessary for reduction by NADPH. | steric role |
| Asp55 | Asp54A | The residue is thought to act with Arg281 within the active site to polarise the phenolic OH bond through hydrogen bonding interactions, activating the oxygen towards conjugate attack at the C4a- hydroperoxyflavin intermediate. These residues, however, do not fulfil the role of a general base towards the phenol substrate. | electrostatic stabiliser |
Chemical Components
aromatic unimolecular elimination by the conjugate base, bimolecular nucleophilic addition, hydride transfer, cofactor used, intermediate formation, proton transfer, bimolecular nucleophilic substitution, overall product formed, unimolecular elimination by the conjugate base, native state of enzyme regeneratedReferences
- Xu D et al. (2001), Biochemistry, 40, 12369-12378. Studies of the Mechanism of Phenol Hydroxylase: Mutants Tyr289Phe, Asp54Asn, and Arg281Met†. DOI:10.1021/bi010962y. PMID:11591156.
- Xu D et al. (2002), Biochemistry, 41, 13627-13636. Studies of the Mechanism of Phenol Hydroxylase: Effect of Mutation of Proline 364 to Serine†. DOI:10.1021/bi020446n. PMID:12427024.
- Enroth C et al. (1998), Structure, 6, 605-617. The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis. DOI:10.1016/s0969-2126(98)00062-8. PMID:9634698.
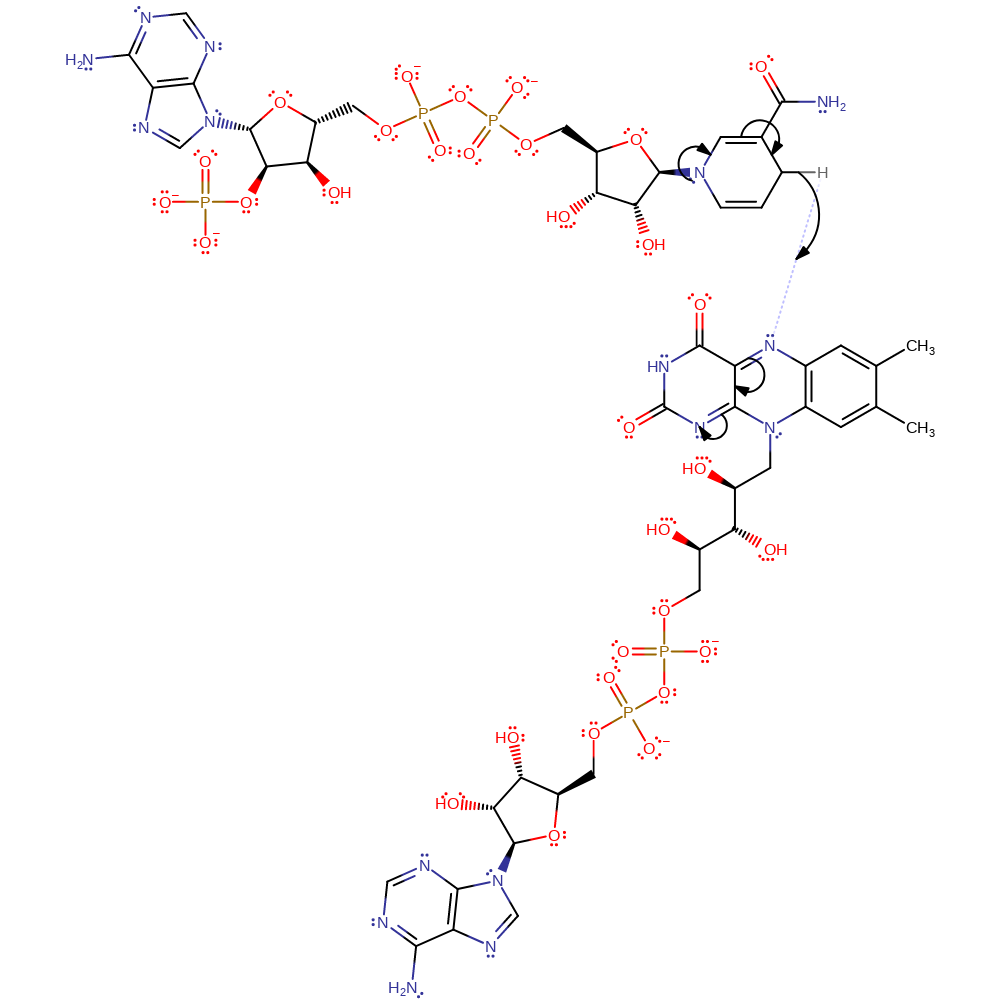
Step 1. In the reductive half reaction a hydride is transferred from NADPH to the flavin cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr289A | steric role |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: bimolecular nucleophilic addition, hydride transfer, cofactor used
Step 2. In the first step of the oxidative half reaction NADP dissociates and the reduced FADH- cofactor immediately reacts with oxygen.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, proton transfer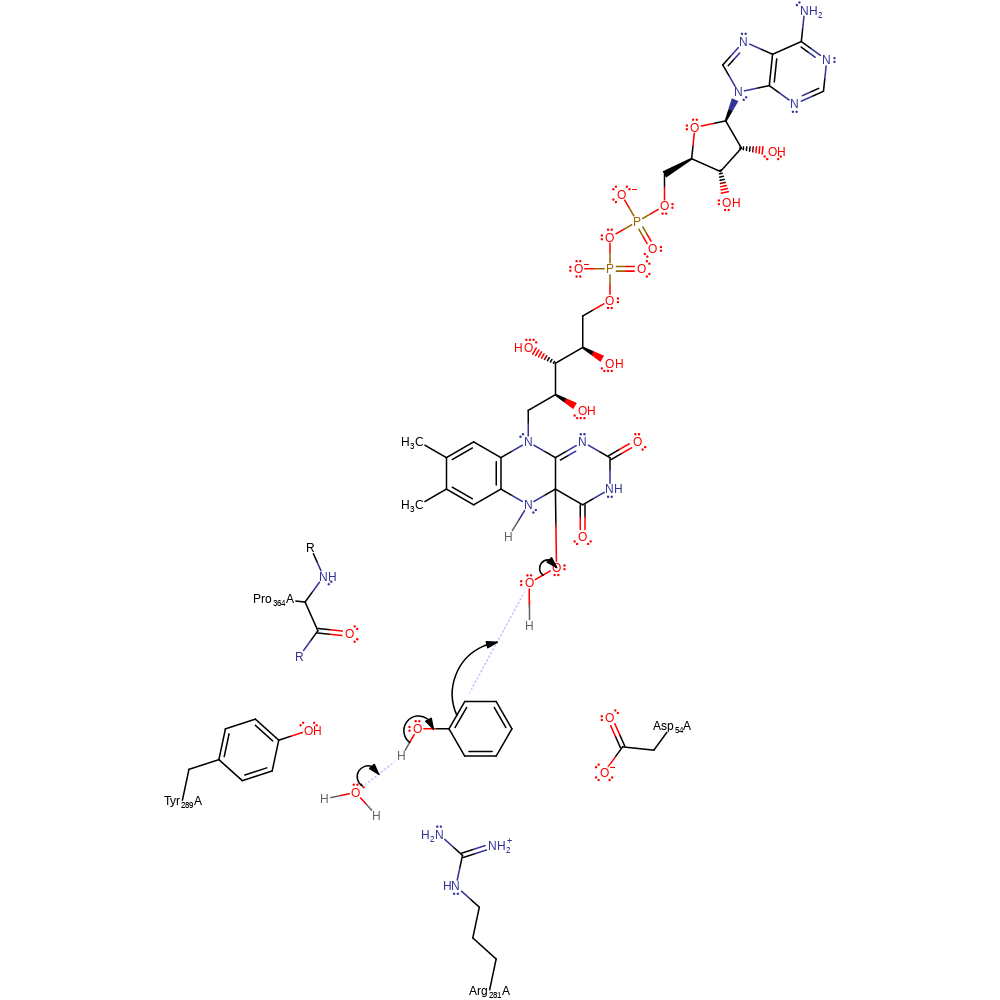
Step 3. Phenol is deprotonated by an unknown base, here represented as water, and initiates nucleophilic attack on the C4a-hydroperoxyflavin intermediate. The resulting C4a-hydroxyflavin is subsequently protonated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp54A | electrostatic stabiliser |
| Arg281A | electrostatic stabiliser |
| Pro364A (main-N) | electrostatic stabiliser, activator |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer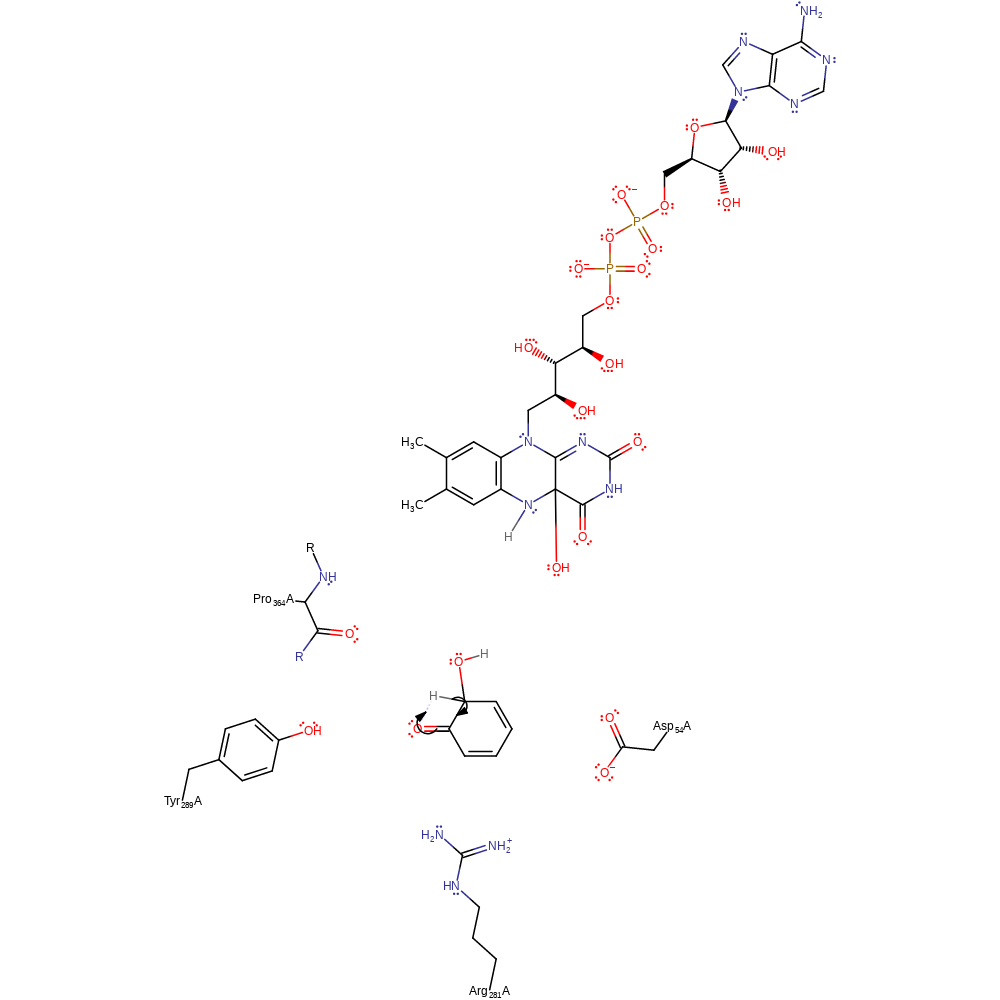
Step 4. Isomerisation of the product to form the catechol product. It is not clear if the proton transfer is aided by other species.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp54A | electrostatic stabiliser |
| Arg281A | electrostatic stabiliser |
| Pro364A (main-N) | electrostatic stabiliser |
Chemical Components
proton transfer, overall product formed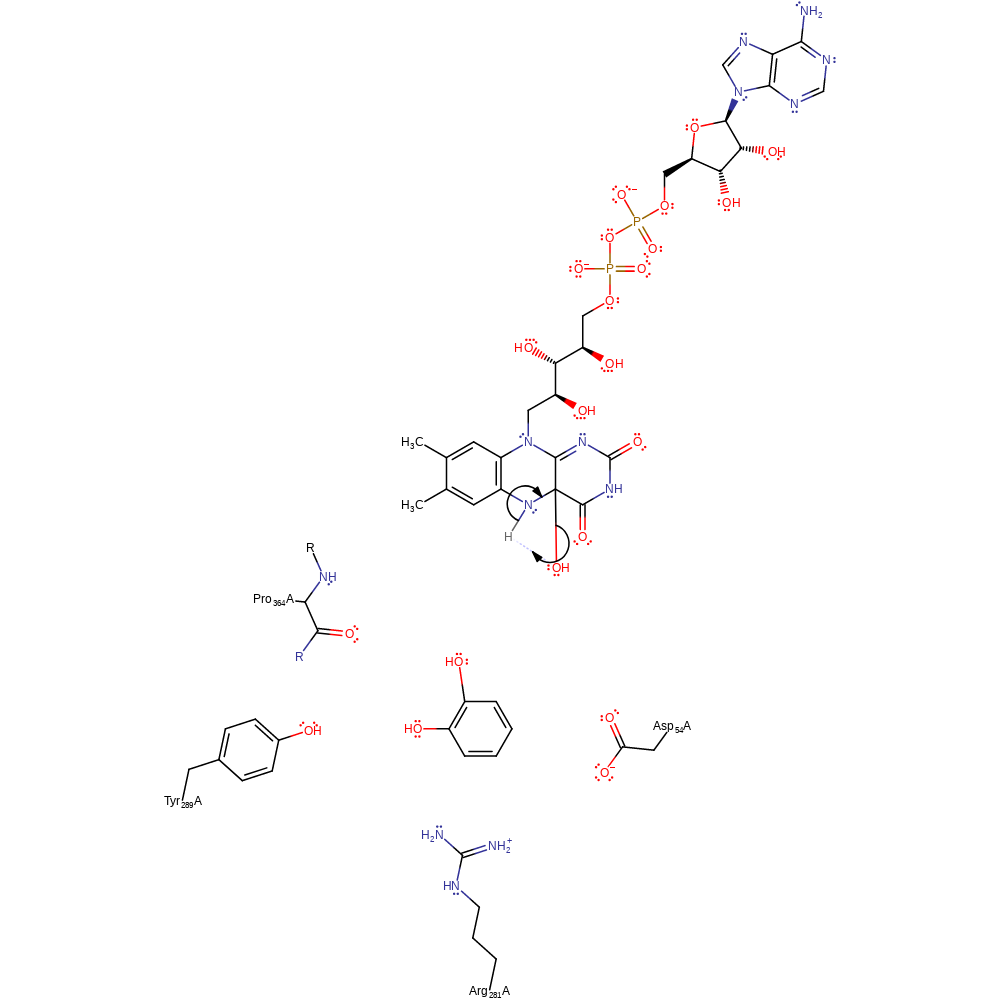
Step 5. The enzyme finishes the reaction cycle by releasing a molecule of water to return the flavin to the oxidized state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|







 Download:
Download: