Endo-1,4-beta-xylanase (glycosyl hydrolase 10)
The Glycosyl hydrolases form a ubiquitous group of enzymes involved in carbohydrate metabolism. This enzyme, from Cellulomonas fimi, belongs to the glucoside hydrolases family (cellulase F) 10.
The class can be sub divided according to the stereoselective reaction outcomes, with hydrolysis occurring with net retention or inversion of the anomeric configuration. This enzyme hydrolyses the beta-glycosidic bond with retention of the anomeric configuration, i.e. it is a retaining glycosidase.
The enzyme has been reported to preferentially cleave a cellobiose unit rather than glucose from the non-reducing end of cellulose, and has been identified as biotechnologically important because of its reaction specificity between xylan and cellulose. It also hydrolyses xylan and, to a lesser extent, carboxymethylcellulose and also a range of soluble aryl xylosides, xylobiosides, glucosides and cellobiosides.
Reference Protein and Structure
- Sequence
-
P07986
 (3.2.1.8, 3.2.1.91)
(3.2.1.8, 3.2.1.91)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Cellulomonas fimi (Bacteria)

- PDB
-
1exp
- BETA-1,4-GLYCANASE CEX-CD
(1.8 Å)



- Catalytic CATH Domains
-
3.20.20.80
 (see all for 1exp)
(see all for 1exp)
Enzyme Reaction (EC:3.2.1.8)
Enzyme Mechanism
Introduction
Retaining glycosidases uses a double-displacement mechanism in which a covalent glycosyl-enzyme intermediate is formed and hydrolysed in a general acid/base catalysed process through oxocarbenium ion-like transition states, or possibly through oxocarbenium ion intermediates.
Cex uses Glu233 as a nucleophile to attack the anomeric carbon in forming the covalent enzyme-substrate intermediate whilst Glu127 acts as the general acid/base to protonate the glycosidic oxygen and activate a water molecule to hydrolyse the enzyme-substrate complex. Transition state stabilisation is achieved by a low barrier hydrogen bond between the sugar C2 hydroxyl group and the carbonyl carbon of the Glu233 carboxylate group.
Catalytic Residues Roles
| UniProt | PDB* (1exp) | ||
| Asn210 | Asn169A | The residue hydrogen bonds to the catalytic Glu 233 residue, activating it for nucleophilic attack at the substrate. | electrostatic stabiliser |
| Glu274 | Glu233A | The residue acts as a nucleophile towards the substrate glycosidic bond, forming an enzyme-substrate intermediate. Interactions though hydrogen bonds with surrounding residues (Asn 169 and His 205) influences the residue's nucleophilicity. | nucleofuge, nucleophile, electrostatic stabiliser |
| Asp276 | Asp235A | The residue's imidazole ring is correctly orientated to hydrogen bond to His 205. This interaction is thought to assist the hydrolysis of the glycosyl-enzyme intermediate by stabilising the released carboxylate residue. | electrostatic stabiliser |
| Glu168 | Glu127A | The residue acts as a general acid towards the breaking glycosidic bond in concert with nucleophilic attack by Glu 233. The residue then acts as a general base towards the hydrolytic water molecule which attacks the enzyme-substrate intermediate. | proton acceptor, proton donor |
| His246 | His205A | The residue hydrogen bonds to the catalytic Glu 233, activating it towards nucleophilic attack at the substrate. | electrostatic stabiliser |
Chemical Components
overall product formed, overall reactant used, enzyme-substrate complex formation, proton transfer, bimolecular nucleophilic substitution, native state of enzyme regenerated, enzyme-substrate complex cleavageReferences
- White A et al. (1996), Nat Struct Biol, 3, 149-154. Crystallographic observation of a covalent catalytic intermediate in a β-glycosidase. DOI:10.1038/nsb0296-149. PMID:8564541.
- Suzuki R et al. (2009), J Biochem, 146, 61-70. Crystallographic Snapshots of an Entire Reaction Cycle for a Retaining Xylanase from Streptomyces olivaceoviridis E-86. DOI:10.1093/jb/mvp047. PMID:19279191.
- Schubot FD et al. (2001), Biochemistry, 40, 12524-12532. Structural basis for the substrate specificity of the feruloyl esterase domain of the cellulosomal xylanase Z from Clostridium thermocellum. PMID:11601976.
- Notenboom V et al. (1998), Nat Struct Biol, 5, 812-818. Insights into transition state stabilization of the β-1,4-glycosidase Cex by covalent intermediate accumulation in active site mutants. DOI:10.1038/1852. PMID:9731776.
- Dominguez R et al. (1995), Nat Struct Biol, 2, 569-576. A common protein fold and similar active site in two distinct families of β-glycanases. DOI:10.1038/nsb0795-569. PMID:7664125.
- MacLeod AM et al. (1994), Biochemistry, 33, 6371-6376. The Acid/Base Catalyst in the Exoglucanase/Xylanase from Cellulomonas fimi Is Glutamic Acid 127: Evidence from Detailed Kinetic Studies of Mutants. DOI:10.1021/bi00186a042. PMID:7910761.
- Hubbard CA et al. (1993), Mov Disord, 8, 473-478. Reversal of reserpine-induced catalepsy by selective D1 and D2 dopamine agonists. DOI:10.1002/mds.870080410. PMID:7901761.
- Tull D et al. (1991), J Biol Chem, 266, 15621-15625. Glutamic acid 274 is the nucleophile in the active site of a "retaining" exoglucanase from Cellulomonas fimi. PMID:1678739.

Step 1. Glu233 initiates a nucleophilic attack on the substrate, forming a enzyme-substrate covalent adduct and one of the sugar products.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu233A | electrostatic stabiliser |
| Asn169A | electrostatic stabiliser |
| His205A | electrostatic stabiliser |
| Asp235A | electrostatic stabiliser |
| Glu127A | proton donor |
| Glu233A | nucleophile |
Chemical Components
overall product formed, overall reactant used, enzyme-substrate complex formation, proton transfer, ingold: bimolecular nucleophilic substitution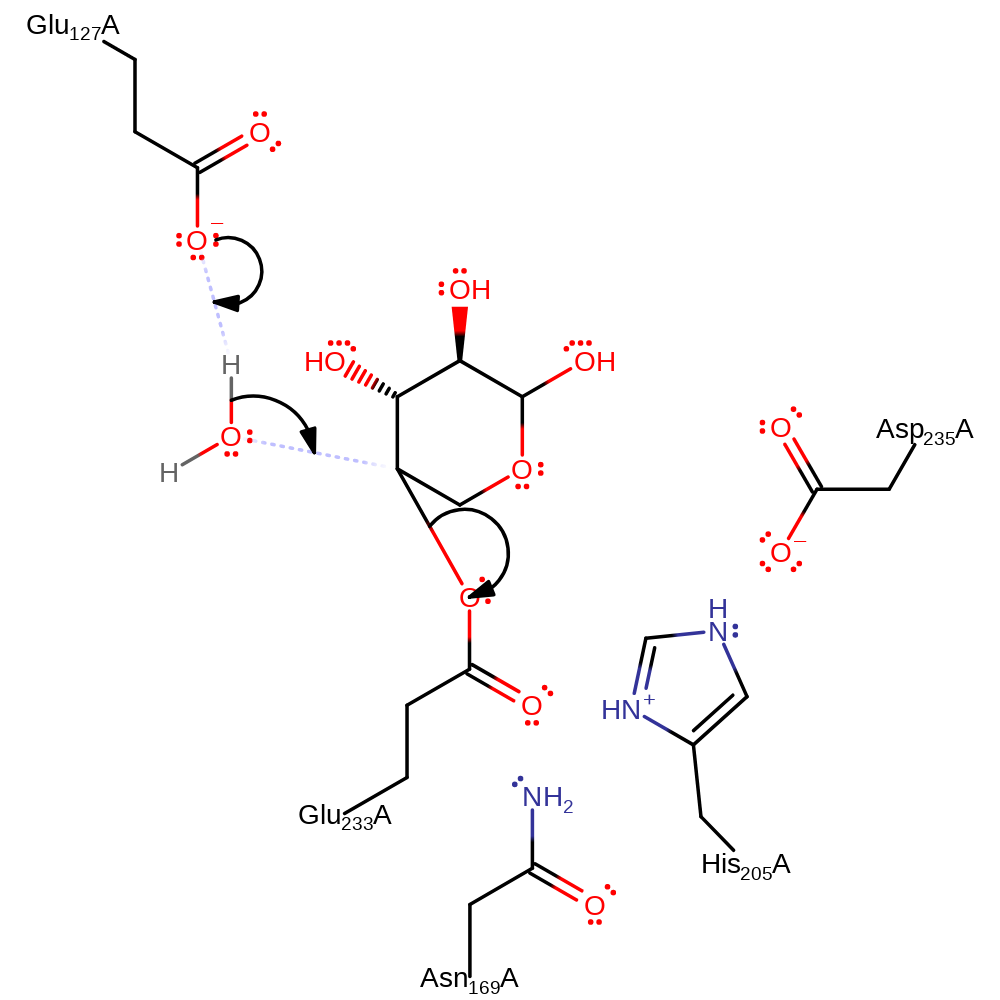
Step 2. Glu127 abstracts a proton from a water molecule, which attacks the intermediate, eliminating Glu233 and forming the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn169A | electrostatic stabiliser |
| His205A | electrostatic stabiliser |
| Asp235A | electrostatic stabiliser |
| Glu233A | nucleofuge |
| Glu127A | proton acceptor |



 Download:
Download: