UDP-N-acetylglucosamine 2-epimerase
The non-hydrolysing bacterial UDP-N-acetylglucosamine 2-epimerase catalyses the conversion of UDP-N-acetylglucosamine to UDP-N-acetylmannosamine, an intermediate in the biosynthesis of several cell surface polysaccarides. The enzyme is allosterically regulated by its substrate, UDP-N-acetylglucosamine, a reaction feature specific to the bacterial form of the enzyme. The structure of the enzyme shows homology with the glycosyl transferases.
Reference Protein and Structure
- Sequence
-
P27828
 (5.1.3.14)
(5.1.3.14)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1f6d
- THE STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE FROM E. COLI.
(2.5 Å)



- Catalytic CATH Domains
-
3.40.50.2000
 (see all for 1f6d)
(see all for 1f6d)
Enzyme Reaction (EC:5.1.3.14)
Enzyme Mechanism
Introduction
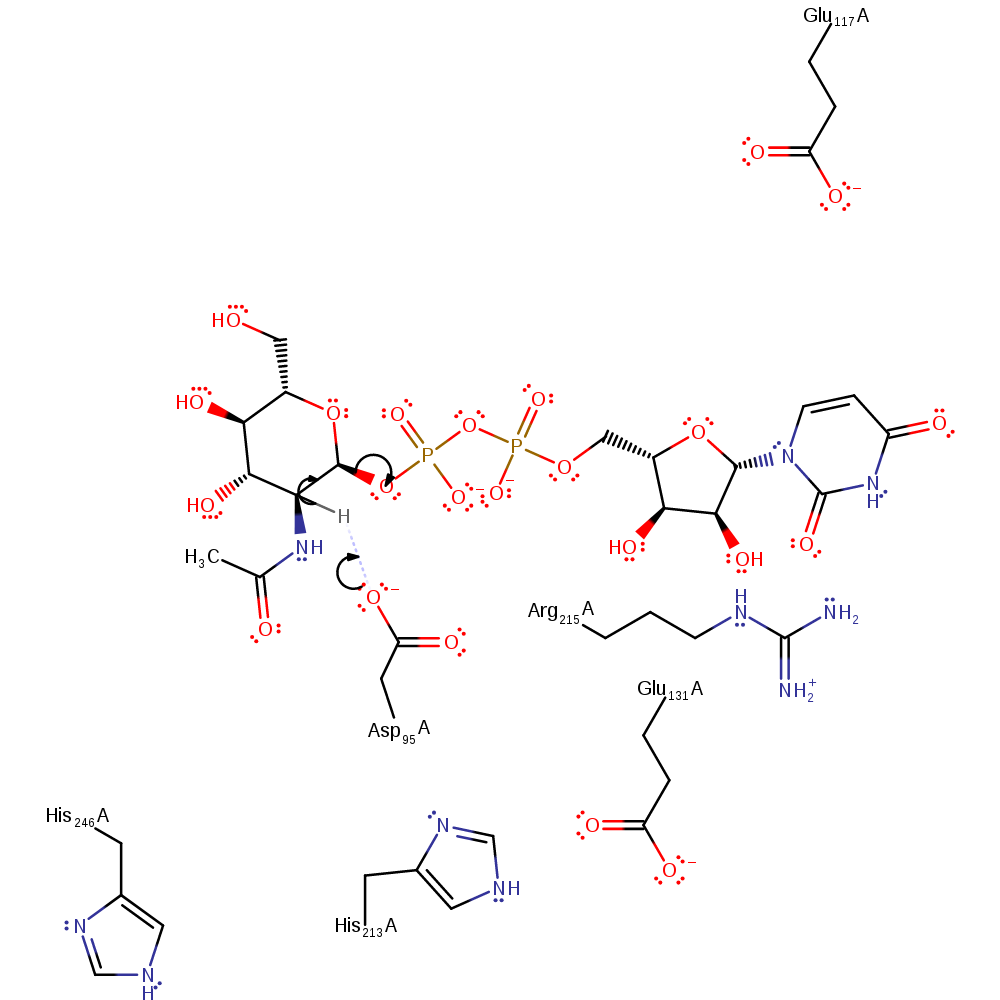
The binding of UDP-N-acetylglucosamine to one subunit induces a conformationa change across the dimer interface that converts the dimer partner to the catalytically active form. It has been suggested that the anit-elimination occurs by an E2 character E1 mechanism, with the transition state showing significant oxocarbenium ion character. His 213 is a good candidate for acting as a general acid to the departing UDP, generating an intermediate 2-acetamidoglucal. The subsequent syn re-addition of UDP gives the UDP-N-acetylmannosamine product.
Catalytic Residues Roles
| UniProt | PDB* (1f6d) | ||
| His213 | His213A | The residue is positioned correctly to act as a general acid to the departing UDP, forming the transient oxocarbenium-character intermediate 2-acetamidoglucal. | proton shuttle (general acid/base) |
| Arg215 | Arg215A | The residue is hydrogen bonded to the UDP-N-acetylglucosamine hydroxyl that interacts with UDP and to one beta phosphate group UDP, and has the role of stabilising the two beta phosphate oxygen atoms of UDP. The lower Hill coefficient associated with the R215A mutant suggests the residue is important in the allosteric regulation of the enzyme. The residue is brought into position by the large realignment of loop between His213-Glu219 on binding of the allosteric regulating UDP-N-acetylglucosamine. Interestingly, Arg215 is only conserved in bacterial UDP-N-acetylglucosamine 2-epimerases. The crucial role of Arg215 in catalysis and conformation is unique to the bacterial non-hydrolysing UDP-N-acetylglucosamine 2-epimerases. |
electrostatic stabiliser |
| His246 | His246A | The residue is thought to play a key role in allosteric activation and to stabilise the UDP intermediate. | electrostatic stabiliser |
| Glu131 | Glu131A | The residue is thought to participate in catalysis by either acting as a general acid/base towards the substrate C2 position, leading to the formation of the oxocarbenium character intermediate or by stabilising the positive charge of this intermediate through hydrogen bonding. The residue has also been implicated in the allosteric activation of the catalytic site. | proton shuttle (general acid/base), electrostatic stabiliser |
| Glu117, Asp95 | Glu117A, Asp95A | The residue is thought to participate in catalysis by either acting as a general acid/base towards the substrate C2 position, leading to the formation of the oxocarbenium character intermediate or by stabilising the positive charge of this intermediate through hydrogen bonding. | proton shuttle (general acid/base), electrostatic stabiliser |
Chemical Components
References
- Velloso LM et al. (2008), EMBO Rep, 9, 199-205. A structural basis for the allosteric regulation of non-hydrolysing UDP-GlcNAc 2-epimerases. DOI:10.1038/sj.embor.7401154. PMID:18188181.
- Chen SC et al. (2016), Sci Rep, 6, 23274-. Mechanism and inhibition of human UDP-GlcNAc 2-epimerase, the key enzyme in sialic acid biosynthesis. DOI:10.1038/srep23274. PMID:26980148.
- Murkin AS et al. (2004), Biochemistry, 43, 14290-14298. Identification and Mechanism of a Bacterial Hydrolyzing UDP-N-Acetylglucosamine 2-Epimerase†. DOI:10.1021/bi048606d. PMID:15518580.
- Samuel J et al. (2004), Biochim Biophys Acta, 1700, 85-91. Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli. DOI:10.1016/j.bbapap.2004.03.017. PMID:15210128.
- Campbell RE et al. (2000), Biochemistry, 39, 14993-15001. The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases. PMID:11106477.
- Sala RF et al. (1996), J Am Chem Soc, 118, 3033-3034. Enzymatic Formation and Release of a Stable Glycal Intermediate: The Mechanism of the Reaction Catalyzed by UDP-N-Acetylglucosamine 2-Epimerase. DOI:10.1021/ja960266z.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| His213A | proton shuttle (general acid/base) |
| Arg215A | electrostatic stabiliser |
| Asp95A | electrostatic stabiliser |
| Glu117A | electrostatic stabiliser, proton shuttle (general acid/base) |
| Glu131A | electrostatic stabiliser, proton shuttle (general acid/base) |
| His246A | electrostatic stabiliser |
| Asp95A | proton acceptor |
Chemical Components
Catalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
Catalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
Catalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
Catalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
Catalytic Residues Roles
| Residue | Roles |
|---|