Fructose-bisphosphate aldolase (Class II)
Fructose-1,6-bisphosphate aldolase (FBP-aldolase, EC 4.1.2.13) participates in two major metabolic pathways. In gluconeogenesis, they catalyse the aldol condensation of a ketose, dihydroxyacetone phosphate (DHAP) and an aldose, glyceraldehyde 3-phosphate (G3P) to form the acylic form of fructose 1,6-bisphosphate (FBP). In glycolysis, they catalyse the reverse cleavage reaction. In each pathway, the enzyme-catalysed reaction represents a distinctive stage, a switch between six and three carbon units. The aldol condensation is a key reaction in synthetic chemistry that aldolases catalyse with exquisite control of the stereochemistry.
Aldolases have been divided into two groups, class I and class II, depending primarily on the reaction mechanism. This annotation refers to class II aldolases, which are alpha/beta(8) barrel structures. The class II enzymes are not found in mammals, and have an absolute requirement for a divalent cation, usually Zn(II), and are activated by monovalent cations.
Reference Protein and Structure
- Sequence
-
P0AB71
 (4.1.2.13)
(4.1.2.13)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1b57
- CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE IN COMPLEX WITH PHOSPHOGLYCOLOHYDROXAMATE
(2.0 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1b57)
(see all for 1b57)
- Cofactors
- Zinc(2+) (2), Sodium(+1) (1), Water (1) Metal MACiE
Enzyme Reaction (EC:4.1.2.13)
Enzyme Mechanism
Introduction
The aldol condensation carried out by FBP aldolases involves three major covalency changes. (1) Abstraction of the 1-proS proton, DHAP C1-H to produce the ene-diolate. (2) Carbon-carbon bond formation to covalently link DHAP C1 with G3P C1 and so form the FBP C3-C4 bond. (3) A second proton transfer converts C4=O to C4-O-H to complete the synthesis of FBP.
A mechanism could be proposed for the class II FBP-aldolase in the condensation direction as follows [PMID:9878448]:
- DHAP binds either with or following monovalent cation binding and chelates to Zn(II) replacing the two water molecule ligands. Asp-109 is protonated by one of these solvents following zinc stimulated activation of water. Direct coordination of DHAP to the metal assists the precise alignment of catalysis allowing zinc to function as a Lewis acid and to polarise the carbonyl bond of the ketose substrate ready for the condensation. The carbonyl group is polarised, increasing the acidity of the hydroxymethylene hydrogen atoms and promoting abstraction of the proton.
- The deprotonation of DHAP leads to carbanion formation and provides the unsaturated linkage where addition occurs. A base is required to abstract the acidic 1-proS alpha-H. The loop carrying Glu-182 undergoes a conformational change to position the residue closer to the catalytic metal, and carries out the proton abstraction.
- The aldehyde (G3P) binds to Arg-331 and is brought into position to interact with the nucleophilic ene-diolate. The two planes of the ene-diolate nucleophile and the carbonyl acceptor must be near parallel in the initial alignment that precedes C-C bond formation. Asp-109 polarises the carbonyl group of G3P C1.
- The C-C bond is formed and proton transfer occurs. Asp-109 transfers a proton to convert C4=O to a hydroxyl group either in concert with or very quickly after C-C bond formation.
- Product release is regulated by the relative concentrations of DHAP, G3P and FBP.
Catalytic Residues Roles
| UniProt | PDB* (1b57) | ||
| Glu183 | Glu182A | It acts as a base to remove the C1-phoS alpha-H of DHAP. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asn287 | Asn286A | It is positioned just below ene-diolate intermediate to stabilise the charged transition state. | hydrogen bond donor, electrostatic stabiliser, steric role |
| Asp110 | Asp109A | It polarises the carbonyl group of G3P C1 to enhance the aldol reaction. It acts as an acid to donate a proton to convert C4=O to a hydroxyl group to yield the product. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| His227, His111, His265 | His226A, His110A, His264A | Forms part of the Zn(II) binding site. | metal ligand |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, overall reactant used, intermediate formation, bimolecular nucleophilic addition, aldol addition, intermediate terminated, overall product formed, rate-determining step, native state of enzyme regenerated, proton relay, inferred reaction step, cyclisation, intramolecular nucleophilic addition, reaction occurs outside the enzymeReferences
- Hall DR et al. (1999), J Mol Biol, 287, 383-394. The crystal structure of Escherichia coli class II fructose-1,6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity11Edited by R. Huber. DOI:10.1006/jmbi.1999.2609. PMID:10080900.
- Shams F et al. (2014), Biochem Soc Trans, 42, 1792-1795. Fructose-1,6-bisphosphate aldolase (FBA)–a conserved glycolytic enzyme with virulence functions in bacteria: ‘ill met by moonlight’. DOI:10.1042/bst20140203. PMID:25399608.
- Macomber L et al. (2011), Mol Microbiol, 82, 1291-1300. Fructose-1,6-bisphosphate aldolase (class II) is the primary site of nickel toxicity in Escherichia coli. DOI:10.1111/j.1365-2958.2011.07891.x. PMID:22014167.
- Daher R et al. (2010), J Med Chem, 53, 7836-7842. Rational Design, Synthesis, and Evaluation of New Selective Inhibitors of Microbial Class II (Zinc Dependent) Fructose Bis-phosphate Aldolases. DOI:10.1021/jm1009814. PMID:20929256.
- Hall DR et al. (2003), Acta Crystallogr D Biol Crystallogr, 59, 611-614. The organization of divalent cations in the active site of cadmiumEscherichia colifructose-1,6-bisphosphate aldolase. DOI:10.1107/s0907444902023661.
- Zgiby SM et al. (2000), Eur J Biochem, 267, 1858-1868. Exploring substrate binding and discrimination in fructose1, 6-bisphosphate and tagatose 1,6-bisphosphate aldolases. PMID:10712619.
- Plater AR et al. (1999), J Mol Biol, 285, 843-855. Conserved residues in the mechanism of the E. coli class II FBP-aldolase. DOI:10.1006/jmbi.1998.2376. PMID:9878448.
- Blom NS et al. (1996), Nat Struct Biol, 3, 856-862. Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase. DOI:10.1038/nsb1096-856. PMID:8836102.
- Cooper SJ et al. (1996), Structure, 4, 1303-1315. The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold. DOI:10.1016/s0969-2126(96)00138-4. PMID:8939754.
- Berry A et al. (1993), FEBS Lett, 318, 11-16. Identification of zinc-binding ligands in the Class II fructose- 1,6-bisphosphate aldolase ofEscherichia coli. DOI:10.1016/0014-5793(93)81317-s.
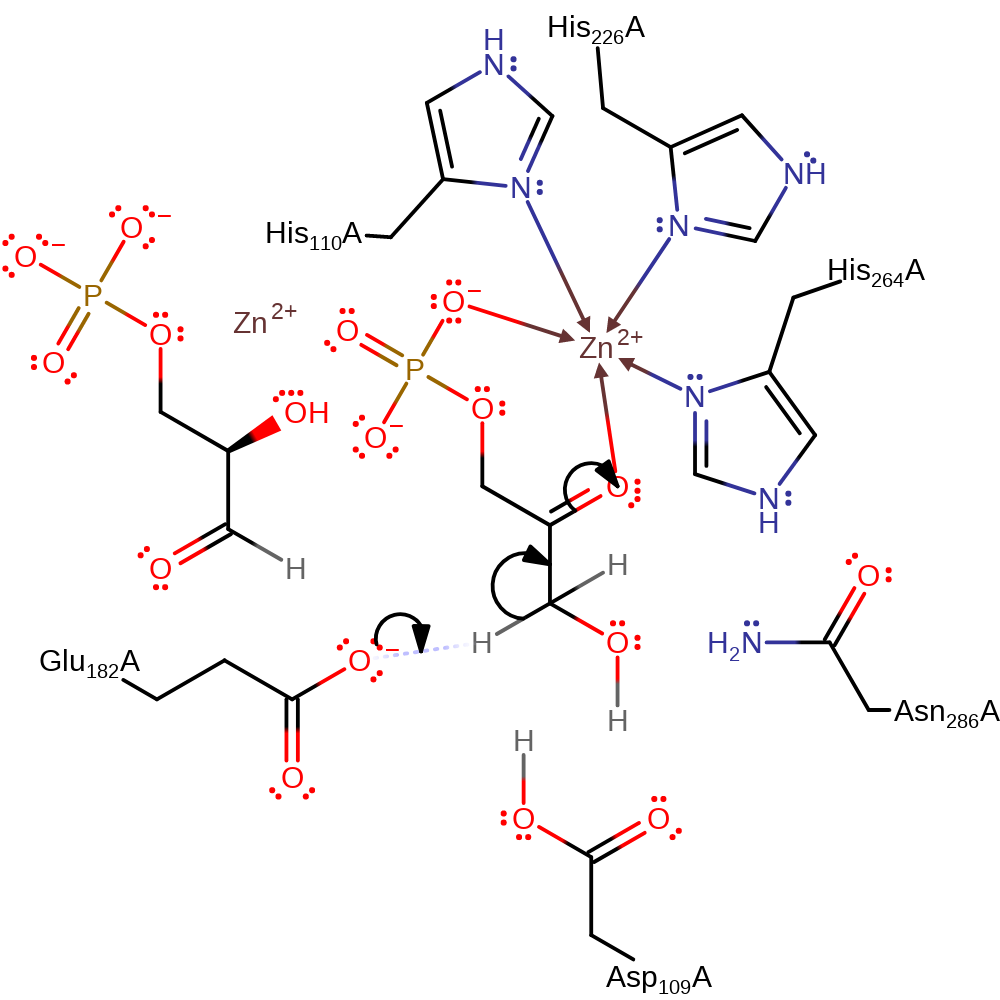
Step 1. Glu182 deprotonates the C1 carbon of the substrate, which initiates double bond rearrangement to form the enol-intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn286A | hydrogen bond donor, electrostatic stabiliser |
| Glu182A | hydrogen bond acceptor |
| Asp109A | hydrogen bond acceptor |
| Glu182A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, overall reactant used, intermediate formation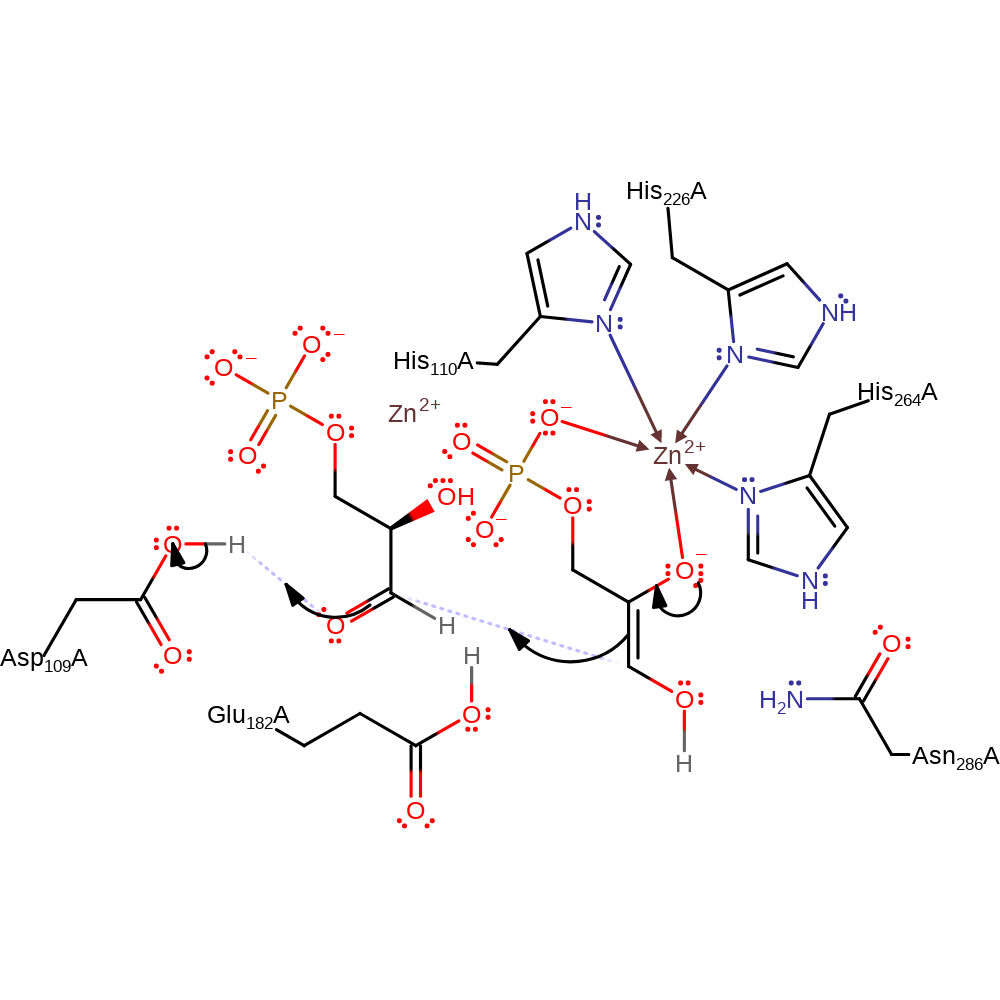
Step 2. The oxyanion collapses, initiating nucleophilic addition of the intermediate to the glyceraldehyde-3-phosphate at the carbonyl carbon. The oxyanion formed deprotonates Asp109.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn286A | electrostatic stabiliser, hydrogen bond donor, steric role |
| Asp109A | hydrogen bond donor, hydrogen bond acceptor |
| Asp109A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, aldol addition, overall reactant used, intermediate terminated, overall product formed, rate-determining step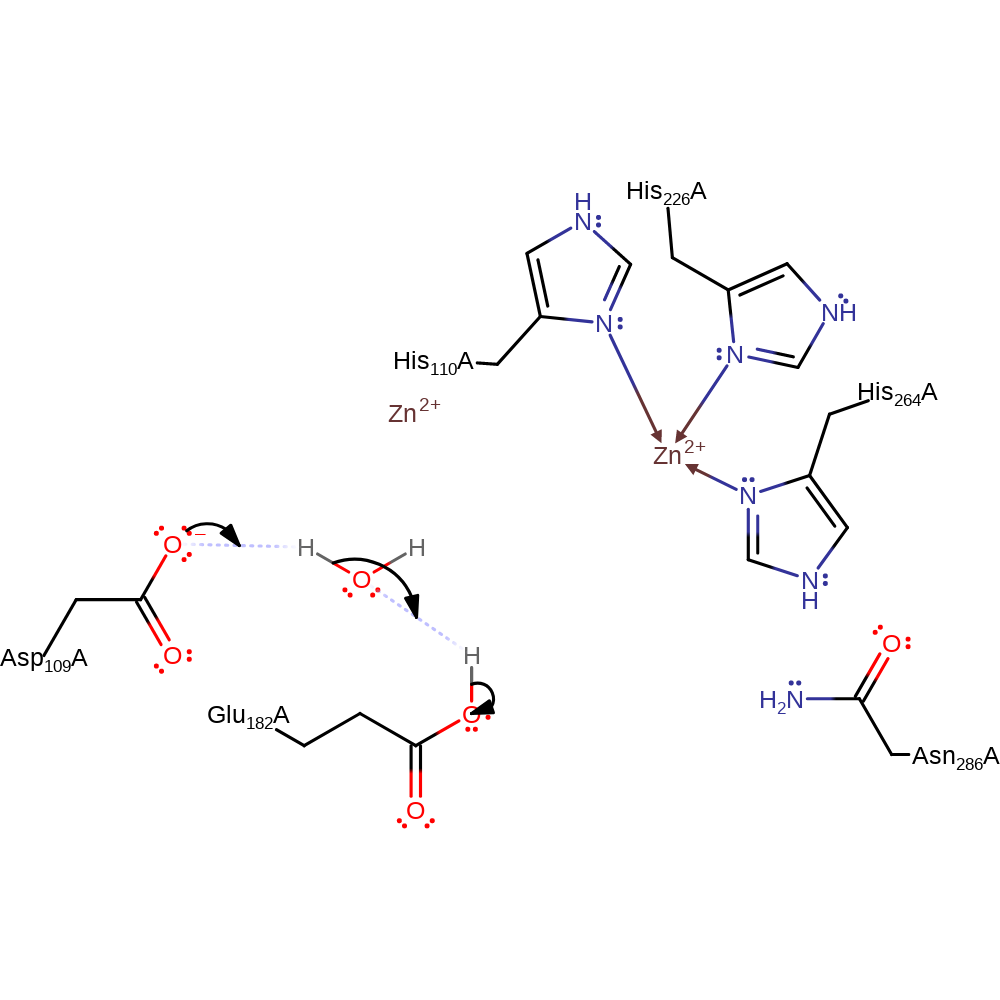
Step 3. Asp109 deprotonates water which deprotonates Glu182 in an inferred step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp109A | hydrogen bond acceptor |
| Glu182A | hydrogen bond donor |
| His226A | metal ligand |
| His110A | metal ligand |
| His264A | metal ligand |
| Asp109A | proton acceptor |
| Glu182A | proton donor |
Chemical Components
proton transfer, native state of enzyme regenerated, proton relay, inferred reaction stepCatalytic Residues Roles
| Residue | Roles |
|---|



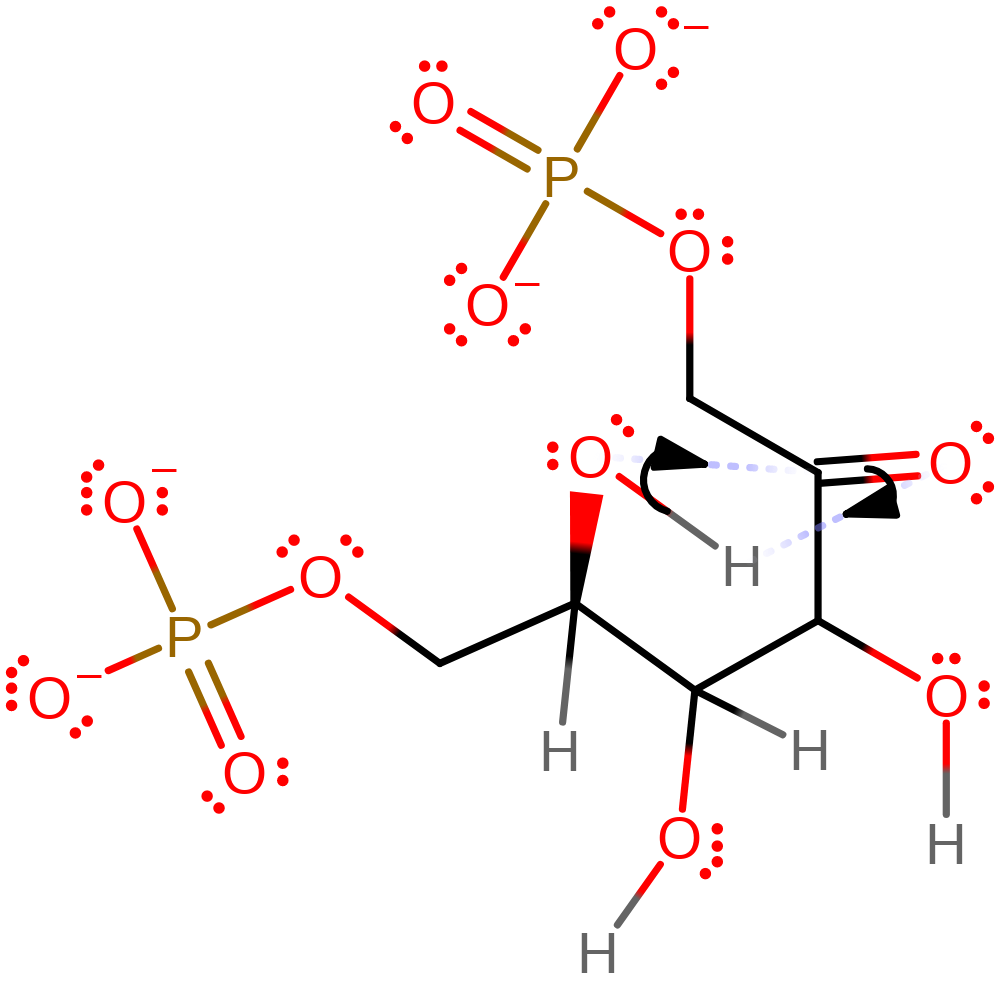
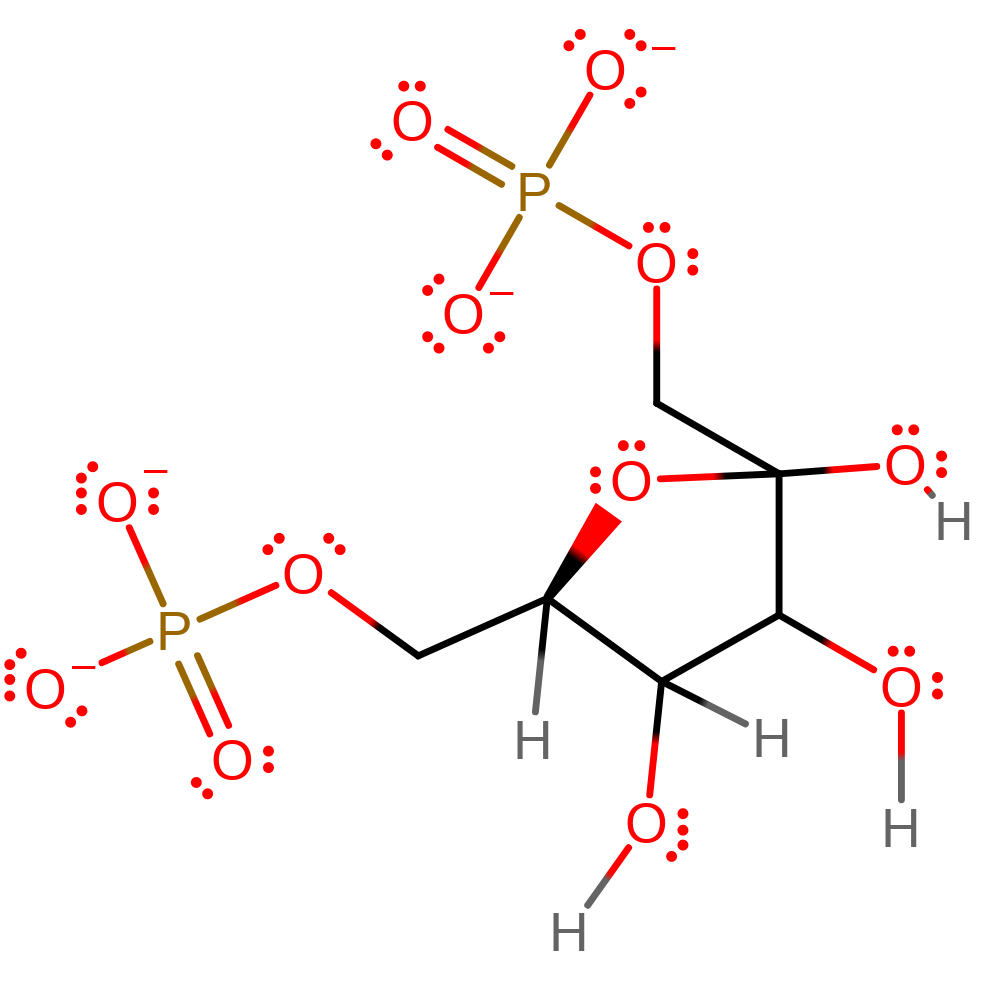 Download:
Download: