3,2-trans-enoyl-CoA isomerase (peroxisomal)
Delta3-delta2-enoyl CoA isomerase (enoyl-CoA isomerase) is a peroxisomal enzyme from the yeast Saccharomyces cerevisiae. It catalyses the conversion of 3-cis-enoyl-CoA or 3-trans-enoyl-CoA into 2-trans-enoyl-CoA. This is involved in an auxiliary pathway of fatty acid degradation, and the product is subsequently metabolised further by other enzymes of the pathway. A yeast strain without the isomerase gene is unable to grow on unsaturated fatty acids, suggesting that this is the only enoyl-CoA isomerase in yeast.
Enoyl-CoA isomerases use fatty acyl substrates with side-chains ranging from C6 to C16. The reaction used as example uses a C12 compund as substrate
Reference Protein and Structure
- Sequence
-
Q05871
 (5.3.3.8)
(5.3.3.8)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1pjh
- Structural studies on delta3-delta2-enoyl-CoA isomerase: the variable mode of assembly of the trimeric disks of the crotonase superfamily
(2.1 Å)



- Catalytic CATH Domains
-
3.90.226.10
 (see all for 1pjh)
(see all for 1pjh)
Enzyme Reaction (EC:5.3.3.8)
Enzyme Mechanism
Introduction
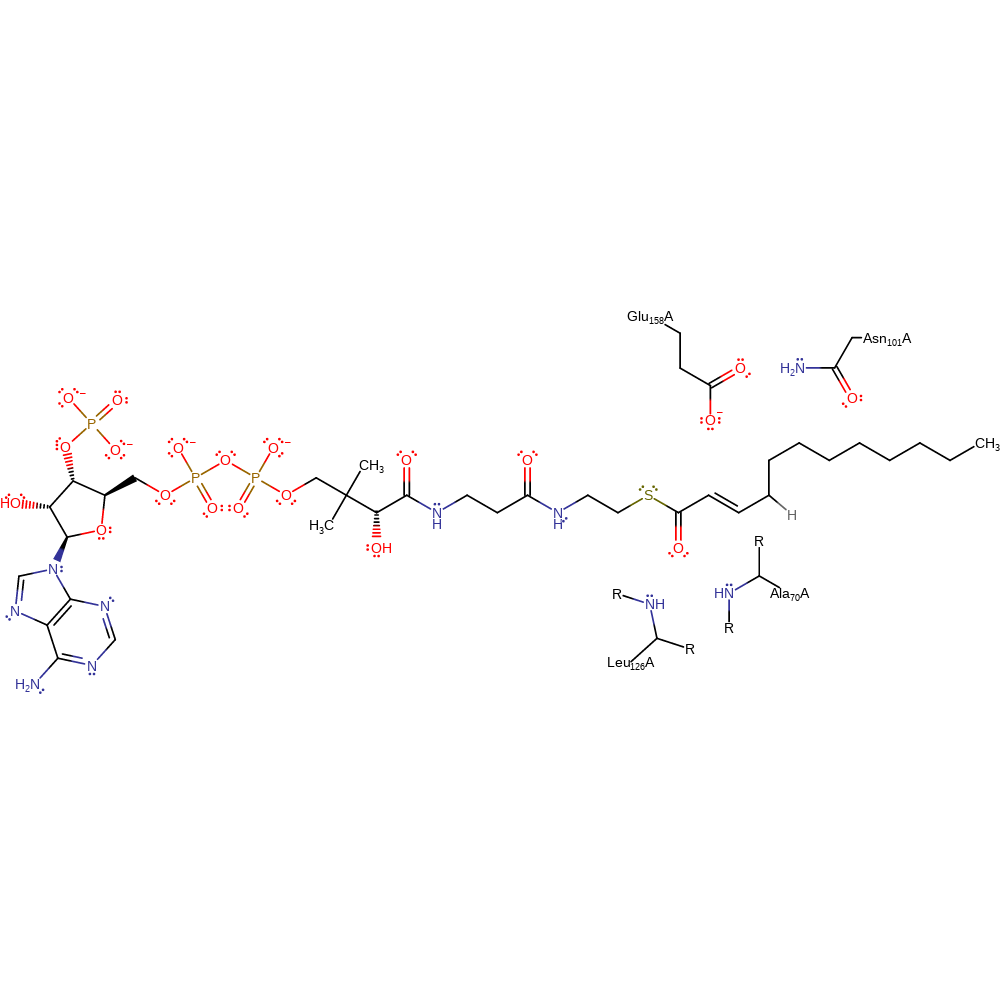
This reaction proceeds via an E1cb mechanism in which Glu158 deprotonates the C2 atom of the substrate, forming the conjugate base. The thioester oxygen atom holds the negative charge, and is stabilised by the presence of an oxyanion hole formed by the amide groups of Ala 70 and Leu 126. As the carbonyl is reformed, the C4 atom of the product is protonated by Glu158.
Catalytic Residues Roles
| UniProt | PDB* (1pjh) | ||
| Glu158 | Glu158A | Deprotonates the C2 atom of the substrate, and donates the proton back to the C4 atom of the product. | proton acceptor, proton donor |
| Ala70 (main-N), Leu126 (main-N) | Ala70A (main-N), Leu126A (main-N) | The amide nitrogen points toward the carbonyl oxygen of the thioester, stabilising the substrate's conjugate base. | electrostatic stabiliser |
| Asn101 | Asn101A | Perturbates the pKa of Glu158 through a hydrogen bonding network, activating the glutamate to act as a general acid/base. | modifies pKa, electrostatic stabiliser |
Chemical Components
assisted keto-enol tautomerisation, proton transfer, assisted tautomerisation (not keto-enol)References
- Mursula AM et al. (2001), J Mol Biol, 309, 845-853. The crystal structure of Δ3-Δ2-enoyl-CoA isomerase. DOI:10.1006/jmbi.2001.4671. PMID:11399063.
- Onwukwe GU et al. (2015), Acta Crystallogr D Biol Crystallogr, 71, 2178-2191. Structures of yeast peroxisomal Δ3,Δ2-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole. DOI:10.1107/s139900471501559x. PMID:26527136.
- Onwukwe GU et al. (2015), FEBS J, 282, 746-768. Human Δ3 ,Δ2 -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10. DOI:10.1111/febs.13179. PMID:25515061.

Step 1. Glu158A deprotonates the substrate with concomitant tautomerisation to the enolate form, which is stabilised by an oxyanion hole composed of the main chain amides of Leu126A and Ala70A
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Leu126A (main-N) | electrostatic stabiliser |
| Ala70A (main-N) | electrostatic stabiliser |
| Asn101A | modifies pKa, electrostatic stabiliser |
| Glu158A | proton acceptor |
Chemical Components
assisted keto-enol tautomerisation, proton transfer
Step 2. The oxyanion intermediate tautomerises to give product with the help of a proton donated by Glu158A.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ala70A (main-N) | electrostatic stabiliser |
| Asn101A | electrostatic stabiliser |
| Leu126A (main-N) | electrostatic stabiliser |
| Asn101A | modifies pKa |
| Glu158A | proton donor |


 Download:
Download: