TRNA pseudouridine38-40 synthase
Pseudouridylate synthase is a member of the pseudouridine synthase family and catalyses the synthesis of pseudourine, the C5-C1' isomer of uridine, in a variety of RNAs. Pseudouridine synthases display varying degrees of specificity for their individual, three dimensionally folded RNA substrates which include tRNA, rRNA and snRNA. Pseudouridine is the most abundant modified nucleoside in the cell and many of the sites targeted for pseudouridine formation, such as those that map to the peptidyl-transfer region of rRNA, are highly conserved in evolution.
Reference Protein and Structure
- Sequence
-
P07649
 (5.4.99.12)
(5.4.99.12)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1dj0
- THE CRYSTAL STRUCTURE OF E. COLI PSEUDOURIDINE SYNTHASE I AT 1.5 ANGSTROM RESOLUTION
(1.5 Å)



- Catalytic CATH Domains
-
3.30.70.660
 3.30.70.580
3.30.70.580  (see all for 1dj0)
(see all for 1dj0)
Enzyme Reaction (EC:5.4.99.12)
Enzyme Mechanism
Introduction
In this proposal Asp60 acts as an acid/base catalyst rather than as a nucleophilic catalyst therefore no enzyme- substrate intermediate is formed. Asp60 facilitates the elimination of the uridine group, which rotates 180 degrees. The uridine group then reattaches to the ribosyl group. See the first proposal for the references.
Catalytic Residues Roles
| UniProt | PDB* (1dj0) | ||
| Asp60 | Asp60(54)A | Act as an acid/base catalyst. | proton acceptor, proton donor |
Chemical Components
overall reactant used, bimolecular elimination, proton transfer, bimolecular nucleophilic addition, assisted keto-enol tautomerisation, overall product formedReferences
- Veerareddygari GR et al. (2016), J Am Chem Soc, 138, 7852-7855. The Pseudouridine Synthases Proceed through a Glycal Intermediate. DOI:10.1021/jacs.6b04491. PMID:27292228.

Step 1. Asp60 acts as a base to facilitate an E2 elimination of the uridine group. This is followed by the 180 degree rotation of the uridine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg205(199)A | electrostatic stabiliser |
| Arg58(52)A | electrostatic stabiliser |
| Asp60(54)A | proton acceptor |
Chemical Components
overall reactant used, ingold: bimolecular elimination, proton transfer
Step 2. The uridine group reattaches to the ribosyl group in a nucleophilic addition reaction. Asp60 now acts as an acid protonating the double bond of the ribosyl group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
| Asp60(54)A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer
Step 3. Asp60 now acting as a base again facilitates the tautomerisation of the substrate into the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
| Asp60(54)A | proton acceptor |
Chemical Components
assisted keto-enol tautomerisation, proton transfer, overall product formedIntroduction
The minimal chemical mechanism for this reaction involves three steps: (i) cleavage of the N-glycosidic linkage of the target uridine (ii) rotation of the uracil ring to position C5 of the pyrimidine near C1' of the ribosyl group of the RNA and (iii) formation of a new C1'-C5 carbon-carbon bond. Asp60 is thought to be catalytic base and is positioned to carry out a nucleophilic attack by electrostatic shielding caused by Arg58 and Arg205 binding specific phosphate groups of the tRNA.
Catalytic Residues Roles
| UniProt | PDB* (1dj0) | ||
| Asp60 | Asp60(54)A | Thought to act as a catalytic nucleophile. | covalently attached, nucleofuge, nucleophile |
| Arg58, Arg205 | Arg58(52)A, Arg205(199)A | Help stabilise the reactive intermediates. | electrostatic stabiliser |
Chemical Components
michael addition, bimolecular nucleophilic addition, overall reactant used, intermediate formation, keto-enol tautomerisation, proton transfer, heterolysis, unimolecular elimination by the conjugate base, overall product formed, intermediate terminated, native state of enzyme regeneratedReferences
- Sivaraman J et al. (2002), Nat Struct Biol, 9, 353-358. Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP. DOI:10.1038/nsb788. PMID:11953756.
- Veerareddygari GR et al. (2016), J Am Chem Soc, 138, 7852-7855. The Pseudouridine Synthases Proceed through a Glycal Intermediate. DOI:10.1021/jacs.6b04491. PMID:27292228.
- Wright JR et al. (2011), RNA, 17, 2074-2084. Pre-steady-state kinetic analysis of the three Escherichia coli pseudouridine synthases TruB, TruA, and RluA reveals uniformly slow catalysis. DOI:10.1261/rna.2905811. PMID:21998096.
- Hur S et al. (2007), Mol Cell, 26, 189-203. How U38, 39, and 40 of Many tRNAs Become the Targets for Pseudouridylation by TruA. DOI:10.1016/j.molcel.2007.02.027. PMID:17466622.
- Hamma T et al. (2006), Chem Biol, 13, 1125-1135. Pseudouridine Synthases. DOI:10.1016/j.chembiol.2006.09.009. PMID:17113994.
- Dong X et al. (2006), RNA Biol, 3, 115-122. Crystal structure of tRNA pseudouridine synthase TruA from Thermus thermophilus HB8. PMID:17114947.
- Ferré-D'Amaré AR (2003), Curr Opin Struct Biol, 13, 683-741. RNA Modifying Enzymes. DOI:10.1016/b978-008045382-8.00671-7. PMID:12581659.
- Mueller EG (2002), Nat Struct Biol, 9, 320-322. Chips off the old block. DOI:10.1038/nsb0502-320. PMID:11976723.
- Foster PG et al. (2000), Nat Struct Biol, 7, 23-27. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. DOI:10.1038/71219. PMID:10625422.
- Ramamurthy V et al. (1999), Biochemistry, 38, 13106-13111. Role of cysteine residues in pseudouridine synthases of different families. PMID:10529181.
- Gu X et al. (1999), Proc Natl Acad Sci U S A, 96, 14270-14275. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. PMID:10588695.
- Huang L et al. (1998), Biochemistry, 37, 344-351. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. DOI:10.1021/bi971874. PMID:9425056.
- Kammen HO et al. (1988), J Biol Chem, 263, 2255-2263. Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. PMID:3276686.

Step 1. Asp60 performs a nucleophilic attack on the tRNA causing an enolate intermediate to be formed in a Michael addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
| Asp60(54)A | covalently attached, nucleophile |
Chemical Components
michael addition, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp60(54)A | covalently attached |
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
Chemical Components
keto-enol tautomerisation, proton transfer
Step 3. The glycosidic bond is cleaved and an oxocarbenium ion is formed. This is followed by a 180 degree rotation of the uridine group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp60(54)A | covalently attached |
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
Chemical Components
heterolysis, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp60(54)A | covalently attached |
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition
Step 5. There is a second keto-enol tautomerisation- the first step in the E1cB elimination of Asp60.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp60(54)A | covalently attached |
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
Chemical Components
proton transfer, keto-enol tautomerisationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
| Asp60(54)A | nucleofuge |
Chemical Components
ingold: unimolecular elimination by the conjugate base, overall product formed, intermediate terminated, native state of enzyme regeneratedIntroduction
In this mechanism Asp60 attaches to the ribosyl group instead of the uridine group in the initial displacement. Similarly to the first proposal there is then a 180 degree rotation of the uridine group. Then finally the reattachment of the urdine group to the ribosyl group with the displacement of Asp60. See the first proposal for the references.
Catalytic Residues Roles
| UniProt | PDB* (1dj0) |
Chemical Components
intermediate formation, overall reactant used, bimolecular nucleophilic substitution, intermediate collapse, overall product formed, proton transfer, tautomerisation (not keto-enol)References
- Gu X et al. (1999), Proc Natl Acad Sci U S A, 96, 14270-14275. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. PMID:10588695.

Step 1. Asp60 attacks the glycosidic bond displacing the uridine group. This is followed by a 180 degree rotation of the uridine group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp60(54)A | covalently attached |
| Arg205(199)A | electrostatic stabiliser |
| Arg58(52)A | electrostatic stabiliser |
| Asp60(54)A | nucleophile |
Chemical Components
intermediate formation, overall reactant used, ingold: bimolecular nucleophilic substitution
Step 2. The rotated uridine group reattaches to the ribosyl group and Asp60 is displaced.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |
| Asp60(54)A | nucleofuge |
Chemical Components
intermediate collapse, ingold: bimolecular nucleophilic substitutionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg58(52)A | electrostatic stabiliser |
| Arg205(199)A | electrostatic stabiliser |


 Download:
Download: 


 Download:
Download: 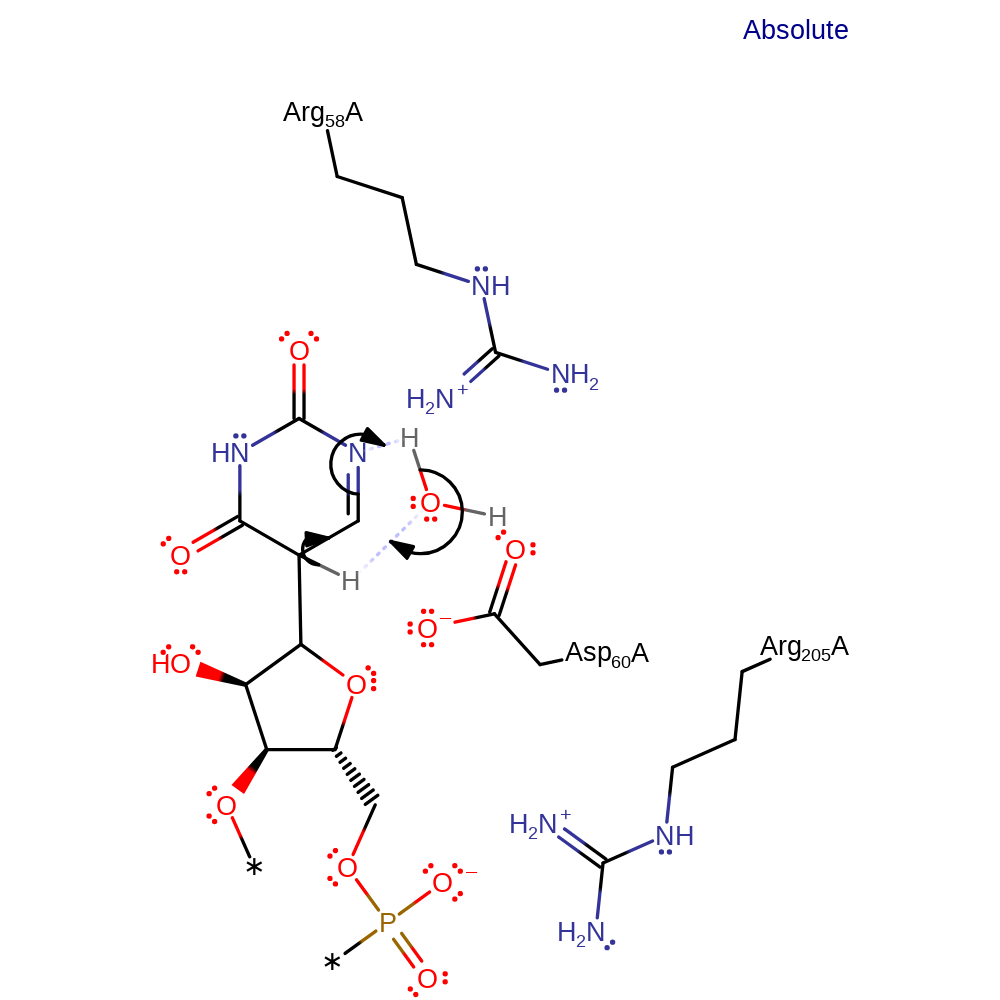
 Download:
Download: