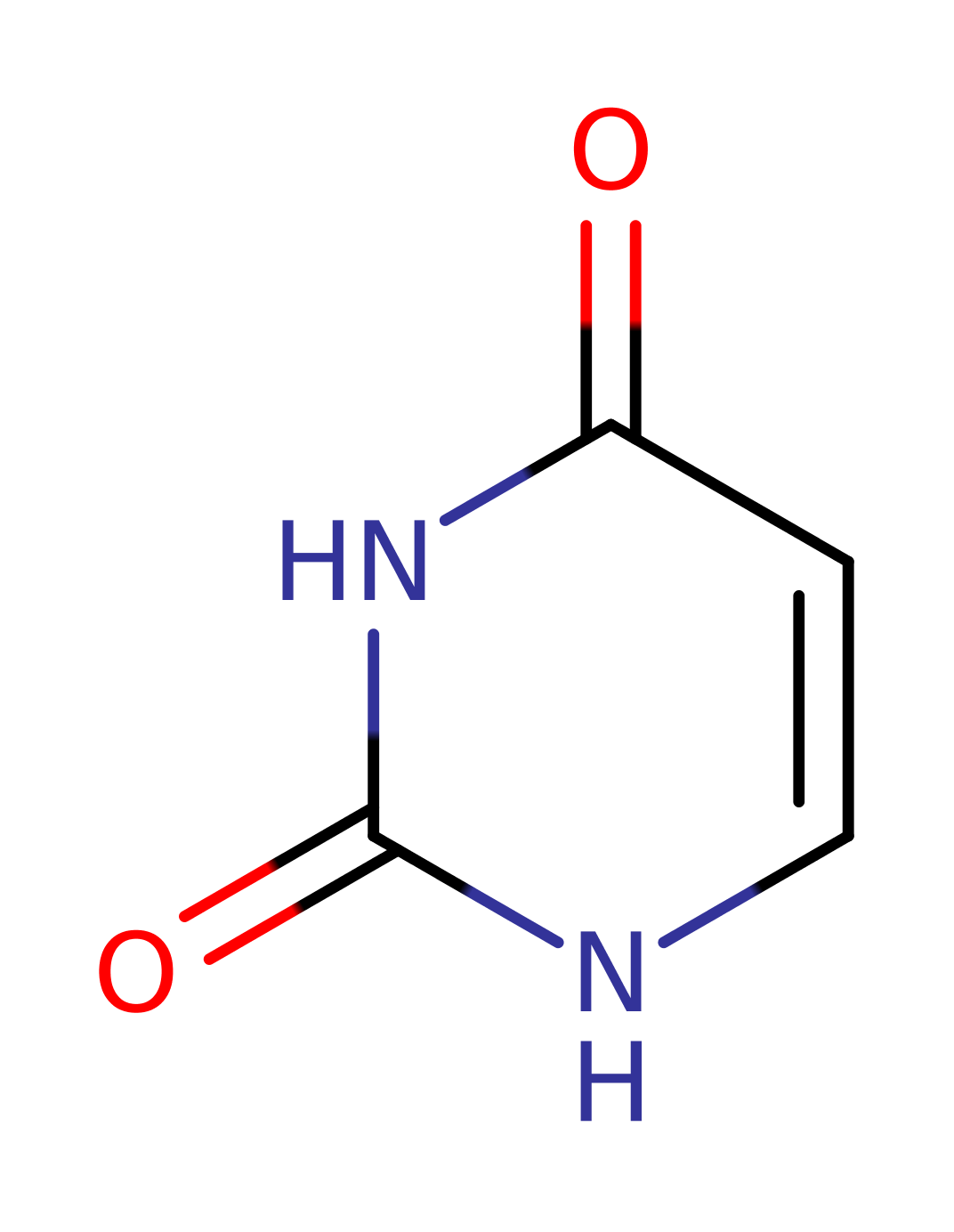
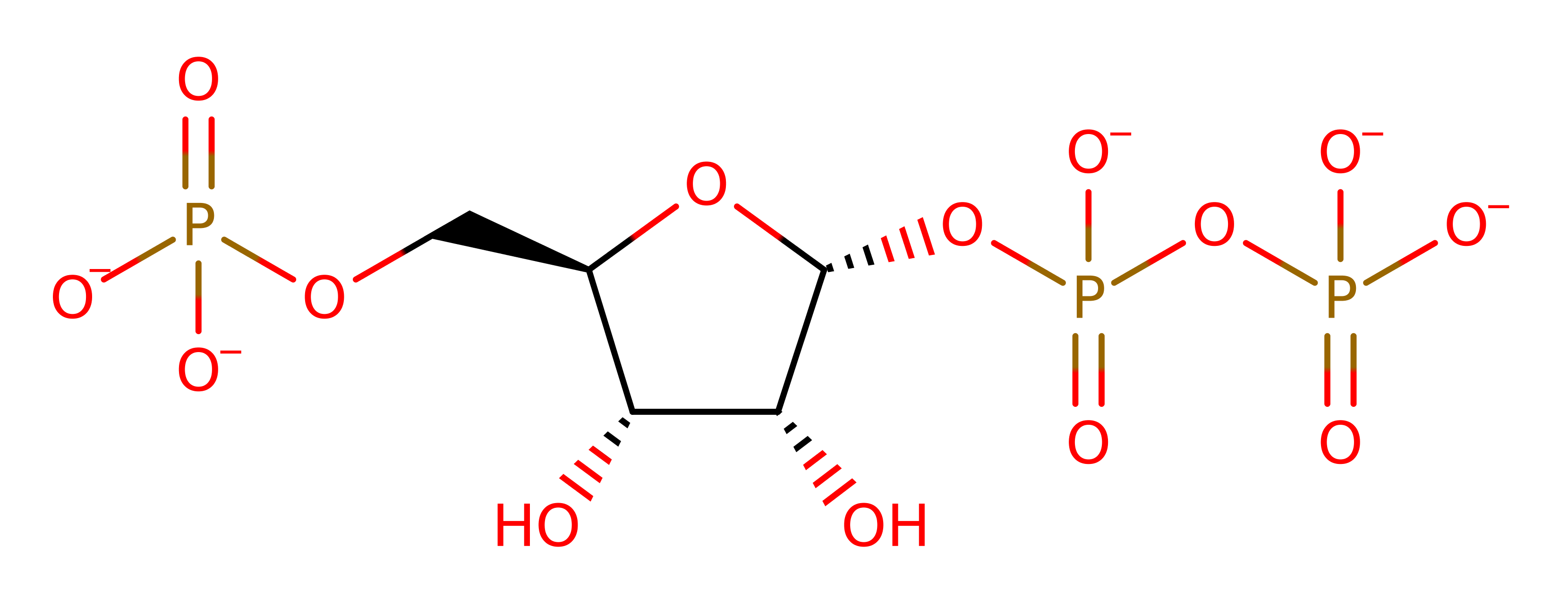
Uracil phosphoribosyltransferase
Catalyses the conversion of uracil and 5-phospho-alpha-D-ribose 1-diphosphate (PRPP) to UMP and diphosphate.
Reference Protein and Structure
- Sequence
-
Q26998
 (2.4.2.9)
(2.4.2.9)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Toxoplasma gondii (Protozoan)

- PDB
-
1bd3
- STRUCTURE OF THE APO URACIL PHOSPHORIBOSYLTRANSFERASE, 2 MUTANT C128V
(1.93 Å)



- Catalytic CATH Domains
-
3.40.50.2020
 (see all for 1bd3)
(see all for 1bd3)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.4.2.9)
+
→
+
Alternative enzyme names: UMP pyrophosphorylase, UMP:pyrophosphate phosphoribosyltransferase, UPRTase, Uridine 5'-phosphate pyrophosphorylase, Uridine monophosphate pyrophosphorylase, Uridylate pyrophosphorylase, Uridylic pyrophosphorylase, UMP diphosphorylase,
Enzyme Mechanism
Introduction
A phosphoribosyl group is transferred from alpha‐d‐5‐phosphoribosyl‐1‐pyrophosphate (PRPP) to the N1 nitrogen of uracil, resulting in the formation of uridine‐5‐monophosphate (UMP) and pyrophosphate (PPi). The inferred transition state is an oxocarbonium species.
Catalytic Residues Roles
| UniProt | PDB* (1bd3) | ||
| Asp238 | Asp238(237)A | Modifies the pKa of Asp235 to ensure it stays in the negative state. | modifies pKa |
| Arg137 | Arg137(136)A | Stabilises the phosphate groups. | electrostatic stabiliser |
| Thr141 | Thr141(140)A | Thr141 is located at the tip of the loop. However, re-positioning of the loop over the active site would allow this residue to enter the catalytic pocket and thus play a role in catalysis. It is likely involved in electrostatic stabilisation. | electrostatic stabiliser |
| Asp235 | Asp235(234)A | Stabilises the transition state. It could also act as a general acid/base with a shift in its side chain position. | electrostatic stabiliser |
*PDB label guide - RESx(y)B(C) - RES: Residue Name; x: Residue ID in PDB file;
y: Residue ID in PDB sequence if different from PDB file; B: PDB Chain;
C: Biological Assembly Chain if different from PDB. If label is "Not Found" it means this residue is not found in the reference PDB.
Chemical Components
References
- Schumacher MA et al. (1998), EMBO J, 17, 3219-3232. Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding. DOI:10.1093/emboj/17.12.3219. PMID:9628859.
- Schumacher MA et al. (2002), Proc Natl Acad Sci U S A, 99, 78-83. The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase. DOI:10.1073/pnas.012399599. PMID:11773618.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp235(234)A | electrostatic stabiliser |
| Arg137(136)A | electrostatic stabiliser |
| Thr141(140)A | electrostatic stabiliser |
| Asp238(237)A | modifies pKa |