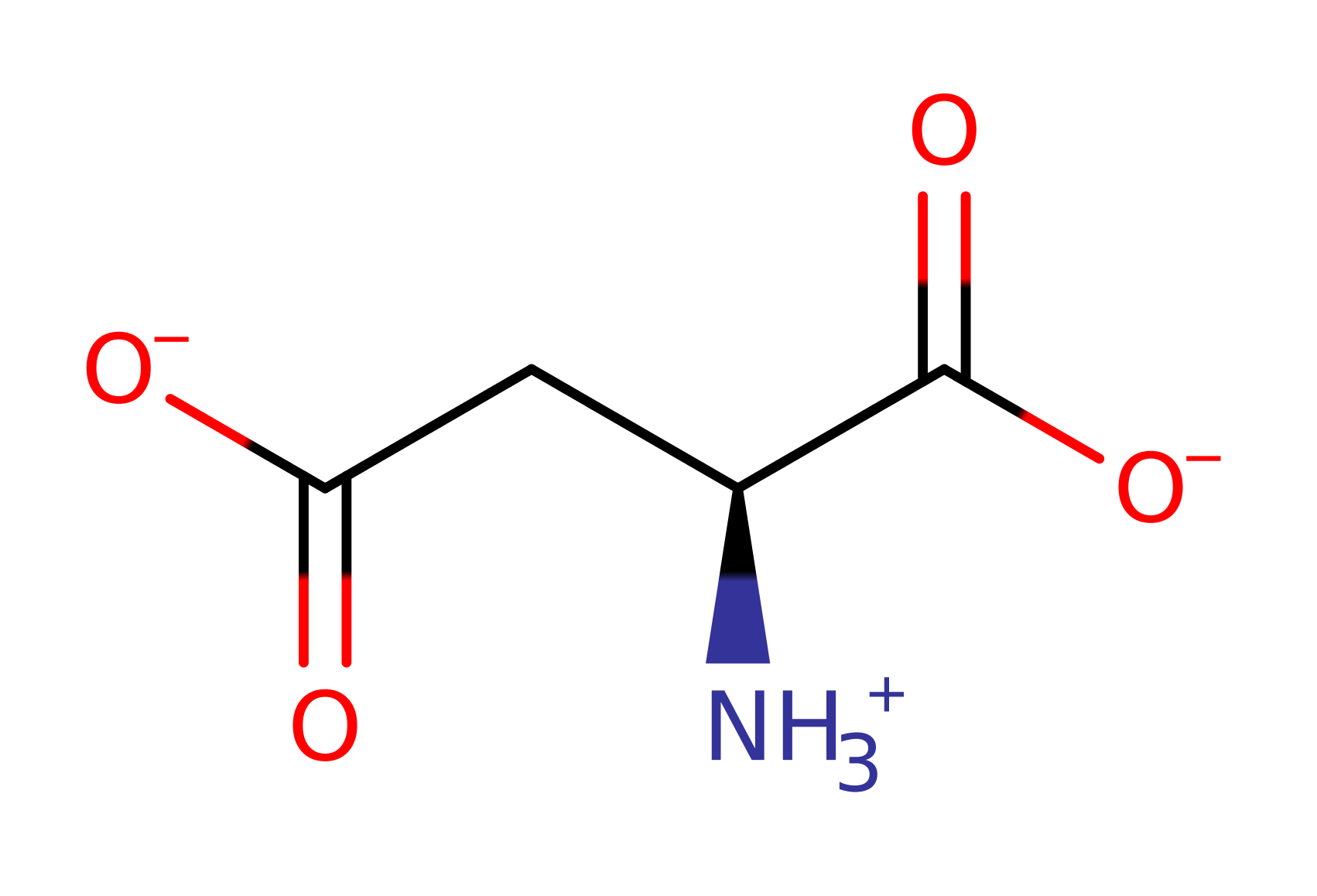
Aspartate---tRNA ligase
Catalyzes the attachment of aspartmate to tRNA(Asp) in a two-step reaction: aspartate is first activated by ATP to form Asp-AMP and then transferred to the acceptor end of tRNA(Asp). Is specific for tRNA(Asp) since it aspartylates tRNA(Asn) 3 orders of magnitude less efficiently than tRNA(Asp).
Aspartyl tRNA synthetase is an alpha2 dimer that belongs to class IIb. Structural analysis combined with mutagenesis and enzymology data on the yeast enzyme point to a tRNA binding process that starts by a recognition event between the tRNA anticodon loop and the synthetase anticodon binding module.
Reference Protein and Structure
- Sequence
-
P04802
 (6.1.1.12)
(6.1.1.12)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1asy
- CLASS II AMINOACYL TRANSFER RNA SYNTHETASES: CRYSTAL STRUCTURE OF YEAST ASPARTYL-TRNA SYNTHETASE COMPLEXED WITH TRNA ASP
(2.9 Å)



- Catalytic CATH Domains
-
3.30.930.10
 (see all for 1asy)
(see all for 1asy)
- Cofactors
- Magnesium(2+) (3)
Enzyme Reaction (EC:6.1.1.12)
Enzyme Mechanism
Introduction
AspRS performs two sequential reactions. The first reaction is the formation of aspartyladenylate from the free amino acid and ATP, releasing pyrophosphate. The second reaction is the displacement of the adenylate moiety by the 3’-OH of the terminal adenosine of the tRNAAsp acceptor arm, yielding the covalently linked Asp-tRNAAsp.
The first reaction occurs through an in-line mechanism with the inversion of configuration at the alpha-phosphorus (the reactive oxygen of the carboxylate group of aspartic acid is located at 2.4 Angstroms from the alpha-phosphate of ATP). This step proceeds via a trigonal bipyramidal pentacovalent intermediate, with the entering and leaving groups occupying apical positions. Stabilization of this reaction intermediate requires a decrease in the negative charge of the alpha-phosphate. The positive charges carried by the guanidium groups of Arg333, Arg325 and Arg531, the imidazole ring of His334 and the magnesium ions altogether participate in the delocalization of the negative charges of the triphosphate moiety of ATP. The binding of one substrate (ATP-Mg2+ or Asp) orients the side chains of the strictly conserved Arg325 and Ser481 such that their side chains are in positions favouring their interaction with the second substrate. This results in a positive coupling which partly compensates for the electrostatic repulsion between the negative charges of the attacking carboxylate group and the receiving alpha-phosphoryl, thereby preparing the catalytic step and stabilising the transition states.
Catalytic Residues Roles
| UniProt | PDB* (1asy) | ||
| Arg325 | Arg325(258)A(C) | Binds and stabilises the negative charge on the alpha-phosphate and substrate aspartate. | electrostatic stabiliser |
| Glu327, Glu478 | Glu327(260)A(C), Glu478(411)A(C) | Binds and stabilises the Mg(II) ions. | metal ligand, electrostatic stabiliser |
| Ser481 | Ser481(414)A(C) | Binds one of the catalytic Mg(II) ions and the substrate aspartate, helping to stabilise the negative charge build-up in the active site. | metal ligand, electrostatic stabiliser |
| Arg531, Arg333, His334 | Arg531(464)A(C), Arg333(266)A(C), His334(267)A(C) | Aids in positioning and stabilising the gamma-phosphate group. | electrostatic stabiliser, steric role |
Chemical Components
References
- Schmitt E et al. (1998), EMBO J, 17, 5227-5237. Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation. DOI:10.1093/emboj/17.17.5227. PMID:9724658.
- Banik SD et al. (2012), J Biomol Struct Dyn, 30, 701-715. Mechanism of the activation step of the aminoacylation reaction: a significant difference between class I and class II synthetases. DOI:10.1080/07391102.2012.689701. PMID:22731388.
- Thompson D et al. (2008), Proteins, 71, 1450-1460. Probing electrostatic interactions and ligand binding in aspartyl-tRNA synthetase through site-directed mutagenesis and computer simulations. DOI:10.1002/prot.21834. PMID:18076053.
- Thompson D et al. (2007), J Biol Chem, 282, 30856-30868. Ammonium Scanning in an Enzyme Active Site: THE CHIRAL SPECIFICITY OF ASPARTYL-tRNA SYNTHETASE. DOI:10.1074/jbc.m704788200. PMID:17690095.
- Thompson D et al. (2006), J Biol Chem, 281, 23792-23803. Molecular Dynamics Simulations Show That Bound Mg2+ Contributes to Amino Acid and Aminoacyl Adenylate Binding Specificity in Aspartyl-tRNA Synthetase through Long Range Electrostatic Interactions. DOI:10.1074/jbc.m602870200. PMID:16774919.
- Moulinier L et al. (2001), EMBO J, 20, 5290-5301. The structure of an AspRS–tRNAAsp complex reveals a tRNA-dependent control mechanism. DOI:10.1093/emboj/20.18.5290. PMID:11566892.
- Sauter C et al. (2000), J Mol Biol, 299, 1313-1324. The free yeast aspartyl-tRNA synthetase differs from the tRNAAsp-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain. DOI:10.1006/jmbi.2000.3791. PMID:10873455.
- Ruff M et al. (1991), Science, 252, 1682-1689. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). DOI:10.1126/science.2047877. PMID:2047877.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg531(464)A(C) | electrostatic stabiliser, steric role |
| Arg333(266)A(C) | electrostatic stabiliser, steric role |
| His334(267)A(C) | electrostatic stabiliser, steric role |
| Arg325(258)A(C) | electrostatic stabiliser |
| Ser481(414)A(C) | metal ligand |
| Glu478(411)A(C) | metal ligand |
| Glu327(260)A(C) | metal ligand |
| Ser481(414)A(C) | electrostatic stabiliser |
| Glu327(260)A(C) | electrostatic stabiliser |
| Glu478(411)A(C) | electrostatic stabiliser |